Deoxyribonucleic acid
Deoxyribonucleic acid, known by the acronym DNA, is a nucleic acid that contains the genetic instructions used in the development and functioning of all living organisms and some viruses. (DNA viruses); it is also responsible for hereditary transmission. The main function of the DNA molecule is the long-term storage of information to build other cell components, such as proteins and RNA molecules. The segments of DNA that carry this genetic information are called genes, but the other DNA sequences have structural purposes or take part in regulating the use of this genetic information.
From a chemical point of view, DNA is a polymer of nucleotides, that is, a polynucleotide. Each nucleotide, in turn, is made up of a carbohydrate (deoxyribose), a nitrogenous base (which can be adenine →A, thymine→T, cytosine→C or guanine→G) and a phosphate group (derived from phosphoric acid). What distinguishes one polynucleotide from another is, then, the nitrogenous base, and therefore the DNA sequence is specified by naming only the sequence of its bases. The sequential arrangement of these four bases along the chain is what encodes the genetic information, following the following complementarity criteria: A-T and G-C. This is because adenine and guanine are larger than thymine and cytosine, so this criterion allows uniformity to be met. In living organisms, DNA occurs as a double strand of nucleotides, in which the two strands are linked together by connections called hydrogen bonds.
In order for the information contained in DNA to be used by the cellular machinery, it must first be copied into shorter trains of nucleotides with different units, called RNA. RNA molecules are copied exactly from DNA by a process called transcription. Once processed in the cell nucleus, the RNA molecules can leave the cytoplasm for later use. The information contained in RNA is interpreted using the genetic code, which specifies the sequence of amino acids in proteins, according to a correspondence of a triplet of nucleotides (codon) for each amino acid. That is, the genetic information (essentially: what proteins are going to be produced at each moment in the life cycle of a cell) is encoded in the nucleotide sequences of DNA and must be translated in order to function. Such translation is done using the genetic code as a dictionary. The "nucleotide sequence-amino acid sequence" allows the assembly of long chains of amino acids (proteins) in the cytoplasm of the cell. For example, in the case of the DNA sequence indicated above (ATGCTAGCATCG...), the DNA polymerase would use the complementary strand of said DNA sequence (which would be TAC-GAT) as a template. -CGT-AGG-...) to transcribe an mRNA molecule that would read AUG-CUA-GCA-UCG-...; the resulting mRNA, using the genetic code, would be translated as the amino acid sequence methionine-leucine-aspartic acid-arginine-...
The DNA sequences that make up the fundamental, physical, and functional unit of heredity are called genes. Each gene contains a part that is transcribed into RNA and another that is responsible for defining when and where they should be expressed. The information contained in genes (genetics) is used to generate RNA and proteins, which are the building blocks of cells, the "bricks" that are used for the construction of organelles or cellular organelles, among other functions.
Inside cells, DNA is organized into structures called chromosomes that, during the cell cycle, are duplicated before the cell divides. Eukaryotic organisms (for example, animals, plants, and fungi) store most of their DNA within the cell nucleus and a minimal part in cellular elements called mitochondria, and in plastids and microtubule organizing centers, or centrioles, if present. have them; prokaryotic organisms (bacteria and archaea) store it in the cytoplasm of the cell and, finally, DNA viruses store it inside the capsid of a protein nature. There are many proteins, such as histones and transcription factors, that bind to DNA, giving it a certain three-dimensional structure and regulating its expression. Transcription factors recognize regulatory sequences in DNA and specify the transcription pattern of genes. The complete genetic material of a chromosome set is called the genome and, with small variations, is characteristic of each species.
History
DNA was first isolated during 1869, by Swiss physician Friedrich Miescher, while working at the University of Tübingen. Miescher was conducting experiments on the chemical composition of pus from discarded surgical bandages when he noticed a precipitate of an unknown substance that he later characterized chemically. He named it nuclein, because he had extracted it from cell nuclei. Nearly 70 years of research were needed to identify the components and structure of nucleic acids.
In 1919 Phoebus Levene identified that a nucleotide is made up of a nitrogenous base, a sugar, and a phosphate. Levene suggested that DNA generated a solenoid (spring)-shaped structure with units of nucleotides linked through the groups phosphate. In 1930 Levene and his teacher Albrecht Kossel proved that Miescher's nuclein is a deoxyribonucleic acid (DNA) made up of four nitrogenous bases (cytosine (C), thymine (T), adenine (A) and guanine (G), the sugar deoxyribose and a phosphate group, and that, in its basic structure, the nucleotide is composed of a sugar attached to the base and phosphate. However, Levene thought that the chain was short and that the bases repeated in a fixed order. In 1937 William Astbury produced the first X-ray diffraction pattern showing that DNA had a regular structure.
The biological function of DNA began to be elucidated in 1928, with a basic series of modern genetics experiments performed by Frederick Griffith, who was working with "smooth" (S) or "rough" (R) strains of the bacterium. Pneumococcus (causing pneumonia), according to the presence (S) or not (R) of a sugary capsule, which is what confers virulence (see also Griffith's experiment). Injecting live S pneumococci into mice kills them, and Griffith observed that if he injected mice with live R pneumococci or heat-killed S pneumococci, the mice did not die. However, if he injected both live R pneumococci and dead S pneumococci, the mice died, and live S pneumococci could be isolated from their blood. Since the dead bacteria could not have multiplied inside the mouse, Griffith reasoned that some kind of change or transformation from one type of bacteria to another must take place by means of a transfer of some active substance, which he called the transforming principle. This substance provided the ability to R pneumococci to produce a sugary capsule and thus become virulent. Over the next 15 years, these initial experiments were replicated by mixing different types of heat-killed and live bacterial strains, both in mice (in vivo) and in test tubes (in vitro). ). The search for the "transforming factor" that was capable of making strains that were not initially virulent continued until 1944, the year in which Oswald Avery, Colin MacLeod, and Maclyn McCarty performed a now-classic experiment. These investigators extracted the active fraction (the transforming factor) and, through chemical, enzymatic, and serological analyses, found that it contained no protein, no unbound lipids, and no active polysaccharides, but consisted mainly of "a viscous form of highly polymerized deoxyribonucleic acid", that is, DNA. DNA extracted from heat-killed S bacterial strains was mixed "in vitro" with live R strains: the result was that S bacterial colonies were formed, thus it was unequivocally concluded that the transforming factor or principle was DNA.
Although the identification of DNA as a transforming principle still took several years to be universally accepted, this discovery was decisive in the knowledge of the molecular basis of heredity, and constitutes the birth of molecular genetics. Finally, the exclusive role of DNA in heritability was confirmed in 1952 by experiments by Alfred Hershey and Martha Chase, in which they found that the T2 phage transmitted its genetic information in its DNA, but not in its protein (see also experiment of Hershey and Chase).
Regarding the chemical characterization of the molecule, in 1940 Chargaff carried out some experiments that helped him to establish the proportions of nitrogenous bases in DNA. He discovered that the ratios of purines were identical to those of pyrimidines, the "equimolecularity" of the bases ([A]=[T], [G]=[C]), and the fact that the amount of G+C in a given DNA molecule is not always equal to the amount of A+T and can vary from 36 to 70 percent of the total content. With all this information and together with X-ray diffraction data provided by Rosalind Franklin, James Watson and Francis Crick proposed the double helix model of DNA in 1953 to represent the three-dimensional structure of the polymer. In a series of five articles in the same issue of Nature, experimental evidence was published that supported the Watson and Crick model. Of these, the article by Franklin and Raymond Gosling was the first publication with X-ray diffraction data supporting the Watson and Crick model, and in that same issue of Nature also featured an article on the structure of DNA by Maurice Wilkins and its his collaborators.
Watson, Crick, and Wilkins were jointly awarded the Nobel Prize in Physiology or Medicine in 1962 after the death of Rosalind Franklin. However, debate continues over who should receive credit for the discovery.
Physical and chemical properties
DNA is a long polymer made up of repeating units, nucleotides. A double strand of DNA is 22 to 26 angstroms (2.2 to 2.6 nanometers) wide, and one unit (a nucleotide) it is 3.3 Å (0.33 nm) long. Although each individual repeating unit is very small, DNA polymers can be huge molecules containing millions of nucleotides. For example, the longest human chromosome, chromosome number 1, is approximately 220 million base pairs long.
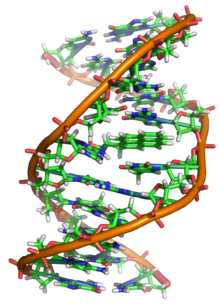
In living organisms, DNA does not usually exist as a single molecule, but rather as a closely associated pair of molecules. The two strands of DNA twist on themselves forming a kind of spiral staircase, called a double helix. The double helix structure model was proposed in 1953 by James Watson and Francis Crick (the article "Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid" was published on April 25, 1953 in Nature), after imaging the double helix structure thanks to X-ray refraction by Rosalind Franklin. The success of this model lay in its consistency with the physical and chemical properties of DNA. The study also showed that the complementarity of bases could be relevant in their replication, and also the importance of the sequence of bases as a carrier of genetic information. Each repeating unit, the nucleotide, contains a segment of the support structure (sugar + phosphate), which holds the strand together, and a base, which interacts with the other DNA strand in the helix. In general, a base attached to a sugar is called a nucleoside, and a base attached to a sugar and one or more phosphate groups is called a nucleotide.
When many nucleotides are linked together, as in DNA, the resulting polymer is called a polynucleotide.
Components
Support structure: The supporting structure of a DNA strand is made up of alternating units of phosphate and sugar (deoxyribose) groups. The sugar in DNA is a pentose, specifically deoxyribose.
- Phosphoric acid:
- Its chemical formula is H3PO4. Each nucleotide may contain one (monophosphate: AMP), two (diphosphate: ADP) or three (triphosphate: ATP) phosphoric acid groups, although as constituent monomers of nucleic acids only appear in the form of monophosphate nucleosides.
- Desoxirribosa:
- It is a monosaccharide of 5 carbon atoms (a pentosa) derived from the ribose, which is part of the nucleotide structure of the DNA. Its formula is C5H10O4. One of the main differences between DNA and RNA is sugar, since in RNA the 2-desoxyrbose of DNA is replaced by an alternative pentase, the ribose.
- Sugar molecules unite with each other through phosphate groups, which form phosphodiéster bonds between third carbon atoms (3′, “three cousins”) and fifth (5′, “five cousin”) of two adjacent sugar rings. Link formation = asymmetrical implies that each strand of DNA has an address. In a double helix, the direction of the nucleotides in a strand (3′ → 5′) is opposed to the direction in the other strand (5′ → 3′). This organization of DNA strands is called anti-parallel; they are parallel chains, but with opposite directions. In the same way, the asymmetric ends of the DNA strands are called extreme 5′ (“five cousin”) and extreme 3′ (“three cousins”), respectively.
- Nitrogenous bases:
- The four majority nitrogenated bases found in DNA are adenine (A), cytosine (C), guanine (G) and thymine (T). Each of these four bases is linked to the sugar-phosphate frame through sugar to form the complete nucleotide (base-sugar-phosphate). The bases are heterocyclic and aromatic compounds with two or more nitrogen atoms, and, within the majority bases, they are classified into two groups: the polymer or purine bases (adenine and guanine), derived from the purine and formed by two rings attached to each other, and the pyramidal bases or pirimidins (cytes) In nucleic acids there is a fifth pirimidynic base, called uracyl (U), which normally occupies the place of thymine in the RNA and differs from that in which it lacks a methyl group in its ring. Uracyl is not usually found in DNA, only rarely appears as a residual product of cytosin degradation due to oxidative desalination processes.
- Timina:
- The genetic code is represented with the letter T. It is a pyramidal derivative with an oxo group in positions 2 and 4, and a methyl group in position 5. It forms the thymidine nucleoside (always desoxitimidine, since it only appears in the DNA) and the thymidylate nucleotide or monophosphate thymidine (dTMP). In DNA, the thymine always pairs with the adenine of the complementary chain through 2 hydrogen bridges, T=A. Its chemical formula is C5H6N2O2 and its nomenclature 2, 4-diox, 5-methylpirimidine.
- Cytosine:
- The genetic code is represented with the letter C. It is a pyramidal derivative, with an amino group in position 4 and an oxo group in position 2. It forms cytidine nucleoside (deoxicitidine in DNA) and cytidilate nucleotide or (desoxi)citidine monophosphate (dCMP in DNA, CMP in RNA). Cytosine is always matched in DNA with the complementary chain guanin by a triple link, C≡G. Its chemical formula is C4H5N3Or and his nomenclature 2-oxo, 4 aminopirimidine. Its molecular mass is 111.10 units of atomic mass. Cytosine was discovered in 1894, insulating it from the tissue of the ram thymus.
- Adenine:
- The genetic code is represented with the letter A. It is a derivative of purine with an amino group in position 6. It forms the adenosine nucleoside (deoxydenosine in DNA) and the adenylate nucleotide or (desoxi)adenosine monophosphate (dAMP, AMP). In DNA it always pairs with the complementary chain timin by 2 hydrogen bridges, A=T. Its chemical formula is C5H5N5 and its nomenclature 6-aminopurine. The adenine, along with the thymine, was discovered in 1885 by the German doctor Albrecht Kossel.
- Guanina:
- The genetic code is represented with the letter G. It is a pubic derivative with an oxo group in position 6 and an amino group in position 2. It forms nucleoside (desoxi)guanosine and the nucleotide guanilato or (desoxi)guanosine monophosphate (dGMP, GMP). The guanine is always paired in DNA with the complementary chain cytosine through three hydrogen links, G≡C. Its chemical formula is C5H5N5Or and his nomenclature 6-oxo, 2-aminopurine.
There are also other nitrogenous bases (the so-called minority nitrogenous bases), derived naturally or synthetically from some other major base. For example, hypoxanthine, relatively abundant in tRNA, or caffeine, both derived from adenine; others, such as acyclovir, derived from guanine, are synthetic analogues used in antiviral therapy; others, such as one of the derivatives of uracil, are antitumor.
Nitrogenous bases have a series of characteristics that give them certain properties. An important characteristic is its aromatic character, a consequence of the presence of double bonds in the conjugated position in the ring. This gives them the ability to absorb light in the ultraviolet part of the spectrum around 260 nm, which can be used to determine the extinction coefficient of DNA and find the existing concentration of nucleic acids. Another of their characteristics is that they present tautomerism or isomerism of functional groups, due to the fact that a hydrogen atom attached to another atom can migrate to a neighboring position; Two types of tautomerism occur in nitrogenous bases: lactam-lactima tautomerism, where hydrogen migrates from nitrogen to oxygen of the oxo group (lactam form) and vice versa (lactam form), and imine-primary amine tautomerism, where hydrogen can be forming the amine group (primary amine form) or migrating to the adjacent nitrogen (imine form). Adenine can only show amine-imine tautomerism, thymine and uracil show double lactam-lactim tautomerism, and guanine and cytosine can show both. On the other hand, and although they are nonpolar molecules, nitrogenous bases have enough polarity to establish hydrogen bonds, since they have highly electronegative atoms (nitrogen and oxygen) that have a partial negative charge, and partially charged hydrogen atoms. positive, so dipoles are formed that allow these weak bonds to form.
The haploid human genome is estimated to be around 3 billion base pairs long. To indicate the size of DNA molecules, the number of base pairs is indicated, and as derivatives there are two widely used units of measurement, the kilobase (kb), which is equivalent to 1000 base pairs, and the megabase (Mb), which equals one million base pairs.
Base Pairing
The DNA double helix is kept stable by the formation of hydrogen bonds between the bases associated with each of the two strands. For the formation of a hydrogen bond, one of the bases must present a "donor" of hydrogens with a partially positively charged hydrogen atom (-NH2 or -NH) and the other base must present an "acceptor" of hydrogens with an electronegatively charged atom (C=O or N). Hydrogen bonds are weaker bonds than typical covalent chemical bonds, such as those that connect the atoms in each strand of DNA, but stronger than individual hydrophobic interactions, Van der Waals bonds, etc. Since hydrogen bonds are not covalent bonds, they can be broken and re-formed relatively easily. For this reason, the two strands of the double helix can be pulled apart like a zipper, either by mechanical force or by high temperature. The double helix is further stabilized by hydrophobic effect and stacking, which are not influenced by the sequence of DNA bases.
Each type of base on one strand forms a bond with only one type of base on the other strand, called base complementarity. Thus, purines form bonds with pyrimidines, such that A binds only to T, and C only to G. The arrangement of two paired nucleotides along the double helix is called base pairing. This pairing corresponds to the observation already made by Erwin Chargaff (1905-2002), who showed that the amount of adenine was very similar to the amount of thymine, and that the amount of cytosine was equal to the amount of guanine in DNA.. As a result of this complementarity, all the information contained in the double-stranded sequence of the DNA helix is duplicated on each strand, which is essential during the DNA replication process. Indeed, this reversible and specific interaction between complementary base pairs is critical for all DNA functions in living organisms.
As noted above, the two types of base pairs form a different number of hydrogen bonds: A=T forms two hydrogen bonds, and C≡G forms three hydrogen bonds (see images). The GC base pair is therefore stronger than the AT base pair. As a consequence, both the percentage of GC base pairs and the total length of the DNA double helix determine the strength of the association between the two DNA strands. Long, high-GC DNA double helices have strands that interact more strongly than short, high-AT double helices. For this reason, areas of the DNA double helix that need to be separated easily tend to have high content in AT, such as the TATAAT sequence of the Pribnow box of some promoters. In the laboratory, the strength of this interaction can be measured by looking for the temperature required to break the hydrogen bonds, the melting temperature (also called the Tm, from English melting temperature). When all the base pairs in a double helix fuse, the strands separate in solution into two completely independent strands. These single-stranded DNA molecules do not have a single common shape, but some conformations are more stable than others.
Other types of base pairs
There are different types of base pairs that can be formed depending on how hydrogen bonds are formed. Those seen in the DNA double helix are called Watson-Crick base pairs, but there are also other possible base pairs, such as Hoogsteen and Wobble u oscillating, which may appear in particular circumstances. In addition, for each type there is the same reverse pair, that is, the one that occurs if the pyrimidine base is rotated 180º on its axis.
- Watson-Crick (base pairs of double helix): the groups of the púrica base that intervene in the hydrogen link are those that correspond to positions 1 and 6 (N acceptr and -NH2 donor if purine is an A) and the groups of the pyramid base, those found in positions 3 and 4 (-NH donator and C=O acceptr if pirimidine is a T). The groups of positions 2 and 3 of the pyramid base (see images) would participate in the reverse Watson-Crick bases.
- Hoogsteen: in this case the groups of the Puric base change, which offers a different face (positions 6 and 7) and which form links with the groups of the pyramids of positions 3 and 4 (as in Watson-Crick). There may also be reverse Hoogsteen. With this type of link you can join A=U (Hoogsteen and Hoogsteen reverse) and A=C (Hoogsteen reverse).
- Wobble or oscillating: this type of link allows you to join guanine and thymine with a double link (G=T). The Puric base (G) forms a link with the groups of positions 1 and 6 (as in Watson-Crick) and the pirimidine (T) with the groups of positions 2 and 3. This type of link would not work with A=C, as the 2 acceptors and the 2 donors would be faced, and could only be given in the reverse case. We found pairs of oscillating-type bases in the RNA, during the mating of codon and anticodon. With this type of link you can join G=U (reverse oscillator) and A=C (reverse oscillator).
In total, in its majority tautomeric form, there are 28 possible pairs of nitrogenous bases: 10 possible purine-pyrimidine base pairs (2 Watson-Crick and 2 reverse Watson Crick pairs, 1 Hoogsteen pair and 2 reverse Hoogsteen pairs, 1 wobble pair and 2 reverse wobble pairs), 7 homopurine-purine pairs (A=A, G=G), 4 A=G pairs, and 7 pyrimidine-pyrimidine pairs. This without counting the pairs of bases that can be formed if we also take into account the other minor tautomeric forms of the nitrogenous bases; these, moreover, may be responsible for transition-type substitution point mutations.
Structure
DNA is a double-stranded molecule, that is, it is made up of two strands arranged antiparallel and with the nitrogenous bases facing each other. In its three-dimensional structure, different levels are distinguished:
- Primary structure
- Sequence of chained nucleotides. It is in these chains where genetic information is found, and since skeleton is the same for all, the difference in information lies in the different sequence of nitrogenated bases. This sequence presents a code, which determines one information or another, according to the order of the bases.
- Secondary structure
- It's a double helix structure. It allows to explain the storage of genetic information and the DNA duplication mechanism. It was postulated by Watson and Crick, based on the X-ray diffraction that Franklin and Wilkins had made, and on the equivalence of Chargaff bases, according to which the sum of more guanine adenins is equal to the sum of more cytosine thymines.
- It is a double chain, dextrogy or levogira, depending on the type of DNA. Both chains are complementary, since the adenine and the guanine of one chain are joined, respectively, to the thymine and cytosine of the other. Both chains are anti-parallel, as the end 3' of one faces the end 5' of the homologa.
- There are three models of DNA. Type B DNA is the most abundant and is the one with the structure described by Watson and Crick.
- Tertiary structure
- It refers to how DNA is stored in a reduced space, to form nucleosomes. It varies according to procariot or eukaryotic organisms:
- In procarotes, DNA folds like a super-hero, usually in circular form and associated with a small amount of protein. The same occurs in cellular orgánulos such as mitochondria and chloroplasts.
- In eukaryotes, given that the amount of DNA of each chromosome is very large, packaging must be more complex and compact; for this it takes the presence of proteins, such as histones and other proteins of a non-Histonic nature (in sperms these proteins are protamines).
- Quaternary structure
- The chromatin present in the nucleus has a thickness of 300 Å, since the chromatin fiber of 100 Å is rolled forming a chromatin fiber of 300 Å. The development of nucleosomes receives the name of solenoid. These solenoids are rolled forming the chromatin of the interphasic nucleus of the eukaryotic cell. When the cell enters division, the DNA is more compacted, thus forming chromosomes.
Double Helix Structures
DNA exists in many conformations. However, only the A-DNA, B-DNA, and Z-DNA conformations have been observed in living organisms. The conformation adopted by DNA depends on its sequence, the amount and direction of supercoiling it exhibits, the presence of chemical modifications in the bases, and the conditions of the solution, such as the concentration of metal and polyamine ions. Of the three conformations, the form "B" it is the most common under conditions existing in cells. The two alternative double helices of DNA differ in their geometry and dimensions.
The shape "A" it is a clockwise rotating spiral, broader than "B", with a shallower, wider minor notch, and a narrower, deeper major notch. The "A" it occurs under non-physiological conditions in dehydrated forms of DNA, while in the cell it can occur in hybrid pairings of DNA-RNA strands, as well as in enzyme-DNA complexes.
Segments of DNA in which the bases have been modified by methylation may undergo further conformational changes and take on the "Z" form. In this case, the strands rotate around the axis of the helix in a left-hand rotating spiral, the opposite of the "B" more frequent. These rare structures can be recognized by specific proteins that bind to Z-DNA and are possibly involved in the regulation of transcription.
Quadruplex structures
At the ends of linear chromosomes are specialized regions of DNA called telomeres. The main function of these regions is to allow the cell to replicate the chromosomal ends using the telomerase enzyme, since the enzymes that replicate the rest of the DNA cannot copy the 3' ends. of chromosomes. These specialized chromosome endings also protect the ends of DNA, and prevent them from being processed by DNA repair systems in the cell as damaged DNA that must be fixed. In human cells, telomeres are long areas of Single-stranded DNA containing a few thousand repeats of a single TTAGGG sequence.
These guanine-rich sequences can stabilize chromosome ends by forming structures of stacked sets of four-base units, instead of the base pairs normally found in other DNA structures. In this case, four guanine bases form flat-topped units that are stacked on top of each other, to form a stable quadruple-G structure. These structures are stabilized by hydrogen bonding between the ends of the bases and chelation of an ionic metal. in the center of each unit of four bases. Other structures can also be formed, with the central set of four bases coming either from a single strand folded around the bases, or from several different parallel strands, so that each contributes a base to the central structure.
In addition to these stacked structures, telomeres also form long loop structures, called telomeric loops or T-loops (T-loops). In this case, the single-stranded DNA coils on itself in a wide circle stabilized by telomere-binding proteins. At the end of the T-loop, the single-stranded telomeric DNA is attached to a region of double-stranded DNA. strand because the telomeric DNA strand disrupts the double helix and pairs to one of the two strands. This triple-stranded structure is called a displacement loop or D-loop (D-loop).
Major and minor clefts
The double helix is a right-handed spiral, that is, each of the nucleotide chains rotates to the right; this can be verified if we look, going from bottom to top, in the direction that the segments of the threads that remain in the foreground follow. If the two strands turn to the right, the double helix is said to be right-handed, and if they turn to the left, left-handed (this shape can appear in alternative helices due to conformational changes in DNA). But in the most common conformation adopted by DNA, the double helix is right-handed, with each base pair rotating about 36º with respect to the previous one.
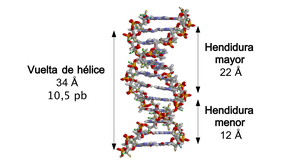
When the two strands of DNA wrap around each other (either to the right or to the left), gaps or slits form between one strand and the other, exposing the sides of the nitrogenous bases inside (see animation).. In the most common conformation that DNA adopts, as a consequence of the angles formed between the sugars of both chains of each pair of nitrogenous bases, two types of clefts appear around the surface of the double helix: one of them, the cleft or major groove, which measures 22 Å (2.2 nm) wide, and the other, the slit or minor groove, which measures 12 Å (1.2 nm) wide. Each turn of the helix, which is when it has made a 360º turn or what is the same, from the beginning of the major cleft to the end of the minor cleft, it will therefore measure 34 Å, and in each of these turns there are about 10.5 bp.
The greater width of the slit implies that the ends of the bases are more accessible in it, so that the number of exposed chemical groups is also greater, which facilitates the differentiation between the base pairs A-T, T-A, C-G, G-C. As a consequence of this, the recognition of DNA sequences by different proteins will also be facilitated without the need to open the double helix. Thus, proteins such as transcription factors that can bind to specific sequences often contact the sides of the bases exposed in the larger cleft. In contrast, the chemical groups that are exposed in the smaller cleft are similar, such that base pair recognition is more difficult; therefore it is said that the larger slit contains more information than the smaller slit.
Sense and antisense
A DNA sequence is called "sense" if its sequence is the same as the sequence of a messenger RNA that is translated into a protein. The sequence of the complementary DNA strand is called "antisense" (antisense). In both DNA strands of the double helix there can be both "sense" sequences, which encode mRNA, and "antisense", which do not. That is, the mRNA-encoding sequences are not all present on just one of the strands, but spread between the two strands. RNAs with antisense sequences are produced in both prokaryotes and eukaryotes, but the function of these RNAs is not entirely clear. It has been proposed that antisense RNAs are involved in the regulation of gene expression through mating. RNA-RNA: Antisense RNAs would pair with complementary mRNAs, thus blocking their translation.
In a few DNA sequences in prokaryotes and eukaryotes —this fact is more frequent in plasmids and viruses—, the distinction between sense and antisense strands is more diffuse, because they have overlapping genes. In these cases, some DNA sequences have a dual function, encoding one protein when read along one strand, and a second protein when read in the opposite direction along the length of the strand. another strand. In bacteria, this overlap may be involved in the regulation of gene transcription, while in viruses overlapping genes increase the amount of information that can be encoded in their tiny genomes.
Supercurl
DNA can be twisted like a rope in a process called DNA supercoiling. When DNA is in a "relaxed" state, a strand normally rotates around the axis of the double helix once every 10.4 base pairs, but if the DNA is twisted the strands may be held together more tightly. or more loosely. If the DNA is twisted in the direction of the helix, supercoiling is said to be positive, and the bases are held together more tightly. If the DNA is twisted in the opposite direction, the supercoiling is called negative, and the bases move away. In nature, most DNA has a slight negative supercoil that is produced by enzymes called topoisomerases. These enzymes are also required to release twisting forces introduced into DNA strands during processes such as transcription and replication.
Chemical Modifications
 |  |  |
| cytosine | 5-methyl-cythosine | timina |
DNA base modifications
Gene expression is influenced by the way DNA is packaged on chromosomes, in a structure called chromatin. Base modifications may be involved in DNA packaging: regions with low or no gene expression usually contain high levels of cytosine base methylation. For example, cytosine methylation produces 5-methyl-cytosine, which is important for X chromosome inactivation. The mean level of methylation varies between organisms: the worm Caenorhabditis elegans lacks cytosine methylation, while vertebrates have a high level—up to 1%—of their DNA contains 5-methyl-cytosine. Despite the importance of 5-methyl-cytosine, it can be deaminated to generate a thymine base. Methylated cytokines are therefore particularly sensitive to mutations. Other base modifications include adenine methylation in bacteria and uracil glycosylation to produce the "base-J" in kinetoplasts.
DNA Damage
DNA can be damaged by many types of mutagens, which change the DNA sequence: alkylating agents, as well as high-energy electromagnetic radiation, such as ultraviolet light and X-rays. The type of damage produced to DNA depends on the type of mutagen. For example, UV light can damage DNA by producing thymine dimers, which are formed by crosslinking between pyrimidine bases. On the other hand, oxidants such as free radicals or hydrogen peroxide produce multiple damages, including base modifications, on all guanine, and double-strand breaks (double-strand breaks). In any given human cell, about 500 bases suffer oxidative damage every day. double-strand breaks, as they are difficult to repair and can cause point mutations, insertions and deletions of the DNA sequence, as well as chromosomal translocations.
Many mutagens are positioned between two adjacent base pairs, which is why they are called intercalating agents. Most intercalating agents are planar aromatic molecules, such as ethidium bromide, daunomycin, doxorubicin, and thalidomide. For an intercalating agent to integrate between two base pairs, they must separate, distorting the DNA strands and opening the double helix. This inhibits transcription and DNA replication, causing toxicity and mutations. For this reason, DNA intercalating agents are often carcinogenic: benzopyrene, acridines, aflatoxin, and ethidium bromide are well-known examples. However, due to their ability to inhibit DNA replication and transcription, these toxins are also used in chemotherapy to inhibit the rapid growth of cancer cells.
DNA damage initiates a response that activates different repair mechanisms that recognize specific DNA lesions, which are repaired on the spot to recover the original DNA sequence. Likewise, DNA damage causes an arrest in the cell cycle, which leads to the alteration of numerous physiological processes, which in turn involves protein synthesis, transport and degradation (see also Checkpoint of DNA damage). Alternatively, if the genomic damage is too great to be repaired, the control mechanisms will induce the activation of a series of cellular pathways that will culminate in cell death.
Biological functions
The biological functions of DNA include information storage (genes and genome), protein coding (transcription and translation), and its self-replication (DNA replication) to ensure the transmission of information to daughter cells during division. cell phone.
Genes and genome
DNA can be considered as a store whose content is the information (message) necessary to build and sustain the organism in which it resides, which is transmitted from generation to generation. The set of information that fulfills this function in a given organism is called the genome, and the DNA that constitutes it, genomic DNA.
Genomic DNA (which is organized into chromatin molecules that in turn assemble into chromosomes) is found in the cell nucleus of eukaryotes, in addition to small amounts in mitochondria and chloroplasts. In prokaryotes, DNA is found in an irregularly shaped body called a nucleoid.
Coding DNA
The information of a genome is contained in the genes, and the set of all the information that corresponds to an organism is called its genotype. A gene is a unit of heredity and is a region of DNA that influences a particular characteristic of an organism (such as eye color, for example). Genes contain an "open reading frame" (open reading frame) that can be transcribed, in addition to regulatory sequences, such as promoters and enhancers, that control the transcription of the open reading frame.
From this point of view, the workers of this mechanism are the proteins. These can be structural, such as muscle proteins, cartilage, hair, etc., or functional, such as hemoglobin or the innumerable enzymes in the body. The main function of heredity is the specification of proteins, DNA being a kind of blueprint or recipe to produce them. Most of the time, the modification of the DNA will cause a protein dysfunction that will give rise to the appearance of some disease. But on certain occasions, the modifications may cause beneficial changes that will give rise to individuals better adapted to their environment.
The approximately thirty thousand different proteins in the human body are made up of twenty different amino acids, and a DNA molecule must specify the sequence in which these amino acids are linked.
In the process of making a protein, the DNA of a gene is read and transcribed into RNA. This RNA serves as a messenger between DNA and the machinery that will make proteins and is therefore called messenger RNA or mRNA. Messenger RNA serves as a blueprint for the machinery that makes proteins, so that it assembles the amino acids in the precise order to assemble the protein.
The central dogma of molecular biology established that the flow of activity and information was: DNA → RNA → protein. However, it has now been shown that this "dogma" It must be expanded, since other information flows have been found: in some organisms (RNA viruses) information flows from RNA to DNA; this process is known as "reverse or reverse transcription", also called "retrotranscription". In addition, it is known that there are DNA sequences that are transcribed into RNA and are functional as such, without ever being translated into protein: they are non-coding RNAs, as is the case with interfering RNAs.
Noncoding DNA
The DNA of an organism's genome can be conceptually divided into two: that which encodes proteins (genes) and that which does not. In many species, only a small fraction of the genome encodes proteins. For example, only about 1.5% of the human genome consists of protein-coding exons (20,000–25,000 genes), while more than 90% consists of non-coding DNA.
Non-coding DNA (also called junk DNA or junk DNA) corresponds to genome sequences that do not generate a protein (from rearrangements, duplications, translocations, and recombinations of viruses, etc.), including introns. Until recently, non-coding DNA was thought to be of no use, but recent studies indicate that this is inaccurate. Among other functions, it is postulated that the so-called "junk DNA" regulates the differential expression of genes. For example, some sequences have affinity for special proteins that have the ability to bind to DNA (such as homeodomains, steroid hormone receptor complexes, etc.), with an important role in the control of transcription and replication mechanisms. These sequences are often called "regulatory sequences," and researchers assume that only a small fraction of those that actually exist have been identified. The presence of both non-coding DNA in eukaryotic genomes and the differences in genome size between species represent a mystery that is known as the "C-value puzzle". Repetitive elements are also functional elements. If they were not considered that way, more than 50% of the total nucleotides would be excluded, since they constitute repeating elements. Recently, a group of researchers from Yale University has discovered a non-coding DNA sequence that would be responsible for human beings having developed the ability to grasp and/or manipulate objects or tools.
On the other hand, some DNA sequences play a structural role in chromosomes: telomeres and centromeres contain few or no protein-coding genes, but are important in stabilizing the structure of chromosomes. Some genes do not code for protein, but are transcribed into RNA: ribosomal RNA, transfer RNA, and interference RNA (RNAi, which are RNAs that block the expression of specific genes). The structure of introns and exons of some genes (such as those for immunoglobulins and protocadherins) are important because they allow for alternative splicing of pre-messenger RNA that makes possible the synthesis of different proteins from the same gene (without this capacity there is no the immune system would exist, for example). Some non-coding DNA sequences represent pseudogenes that have evolutionary value, as they allow the creation of new genes with new functions. Other non-coding DNAs arise from the duplication of small regions of DNA; this is very useful, since the tracing of these repetitive sequences allows phylogenetic studies.
Transcription and translation
In a gene, the sequence of nucleotides along a strand of DNA is transcribed into messenger RNA (mRNA) and this sequence in turn is translated into a protein that an organism is capable of synthesizing or "express" in one or several moments of his life, using the information of said sequence.
The relationship between the nucleotide sequence and the amino acid sequence of the protein is determined by the genetic code, which is used during the process of translation or protein synthesis. The coding unit of the genetic code is a group of three nucleotides (triplet), represented by the initial three letters of nitrogenous bases (eg, ACT, CAG, TTT). The DNA triplets are transcribed into their complementary bases into messenger RNA, and in this case the triplets are called codons (for the example above, UGA, GUC, AAA). In the ribosome, each codon of the messenger RNA interacts with a molecule of transfer RNA (tRNA or tRNA) that contains the complementary triplet, called the anticodon. Each tRNA carries the amino acid corresponding to the codon according to the genetic code, so that the ribosome joins the amino acids to form a new protein according to the "instructions" of the mRNA sequence. There are 64 possible codons, which is why more than one corresponds to each amino acid (due to this duplication of codons, it is said that the genetic code is a degenerate code: it is not univocal); some codons indicate the completion of the synthesis, the end of the coding sequence; these stop codons or stop codons are UAA, UGA and UAG (in English, nonsense codons or stop codons).
DNA Replication
DNA replication is the process by which identical copies or replicas of a DNA molecule are obtained. Replication is essential for the transfer of genetic information from one generation to the next and is therefore the basis of heredity. The mechanism essentially consists of the separation of the two strands of the double helix, which serve as a template for the subsequent synthesis of complementary strands to each of them, which will be called mRNA. The end result is two molecules identical to the original. This type of replication is called semiconservative because each of the two molecules resulting from duplication has a chain from the "mother" and another recently synthesized.
DNA duplication hypothesis
At first, three hypotheses were proposed:
- Semiconservative: According to Meselson-Stahl's experiment, each yarn serves as a mould to form a new yarn, by means of the complentity of bases, with two double helices formed by an old yarn (mould) and a new yarn (copy).
- Conservative: After the doubling the two old strands would remain together and, on the other hand, the two new strands forming a double helix.
- Dispersive: According to this hypothesis, the resulting strands would be made up of fragments in double helice antique DNA and newly synthesized DNA.
DNA-protein interactions
All functions of DNA depend on its interactions with proteins. These interactions may be nonspecific, or the protein may specifically bind to a single DNA sequence. They can also bind enzymes, particularly important among which are polymerases, which copy the base sequences of DNA during transcription and replication.
DNA-binding proteins
Non-specific interactions
 |
Structural proteins that bind to DNA are well-known examples of nonspecific DNA-protein interactions. In chromosomes, DNA is found in complexes with structural proteins. These proteins organize DNA into a compact structure called chromatin. In eukaryotes this structure involves the binding of DNA to a complex formed by small basic proteins called histones, while in prokaryotes a wide variety of proteins are involved. The histones form a cylindrical-shaped complex called nucleosome, around which nearly two turns of double-helical DNA are wound. These non-specific interactions are determined by the existence of basic residues in histones, which form ionic bonds with the sugar-phosphate backbone of DNA and are therefore largely independent of base sequence. These basic amino acids undergo modifications methylation, phosphorylation and acetylation chemistries, which alter the strength of the interaction between DNA and histones, making the DNA more or less accessible to transcription factors and thus modifying the rate of transcription.
Other nonspecific DNA-binding proteins in chromatin include the High Mobility Group (HMG) proteins that bind to DNA. folded or distorted. These proteins are important during the folding of nucleosomes, organizing them into more complex structures to make up chromosomes during the process of chromosome condensation. It has been proposed that other proteins would also intervene in this process, forming a kind of "scaffold" on which chromatin is organized; the main components of this structure would be the enzyme topoisomerase II α (topoIIalpha) and condensin 13S. topoIIalpha in the organization of chromosomes is still disputed, as other groups argue that this enzyme is rapidly exchanged on both chromosome arms and kinetochores during mitosis.
Specific interactions
A well-defined group of DNA-binding proteins is made up of proteins that specifically bind to single-stranded DNA or single-stranded DNA (ssDNA). In humans, replication protein A is the best known of its family and is involved in processes in which the double helix breaks apart, such as DNA replication, recombination, or DNA repair. These proteins appear to stabilize single-stranded DNA., protecting it to prevent it from forming stem-loop structures (stem-loop) or being degraded by nucleases.
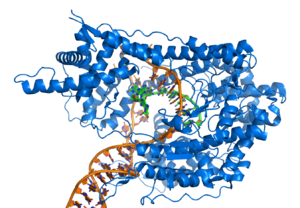
However, other proteins have evolved to specifically bind to particular DNA sequences. The specificity of the interaction of proteins with DNA comes from the multiple contacts with the DNA bases, which allows them to "read" the DNA sequence. Most of these base interactions occur in the larger cleft, where the bases are most accessible.
The specific proteins studied in greater detail are those responsible for regulating transcription, which is why they are called transcription factors. Each transcription factor binds to a specific DNA sequence and activates or inhibits the transcription of genes that have these sequences close to their promoters. Transcription factors can do this in two ways:
- First, they can join the RNA polymerase responsible for transcription, either directly or through other mediating proteins. This way. the union between the polymerase RNA and the promoter is stabilized, allowing the initiation of the transcription.
- Second, transcription factors can join enzymes that modify the promoter's histonas, which alters the accessibility of the DNA mold to the polymerase RNA.
Because target DNAs can be found throughout the genome of the organism, changes in the activity of one type of transcription factor can affect thousands of genes. Consequently, these proteins are often the targets of transduction processes of signals that control responses to environmental changes or cell differentiation and development.
Enzymes that modify DNA
Nucleases and ligases
Nucleases are enzymes that cut DNA strands by catalyzing the hydrolysis of phosphodiester bonds. Nucleases that hydrolyze nucleotides from the ends of DNA strands are called exonucleases, while endonucleases cut inside the strands. The nucleases most frequently used in molecular biology are restriction enzymes, endonucleases that cut DNA at certain specific sequences. For example, the EcoRV enzyme, shown on the left, recognizes the 6-base sequence 5′-GAT|ATC-3′, and cuts both strands at the indicated vertical line, generating two DNA molecules with the blunt ends. Other restriction enzymes, however, generate sticky ends, since they cut the two DNA strands differently. In nature, these enzymes protect bacteria against phage infections by digesting phage DNA as it enters through the bacterial wall, acting as a defense mechanism. In biotechnology, these sequence-specific nucleases DNA is used in genetic engineering to clone fragments of DNA and in the genetic fingerprinting technique.
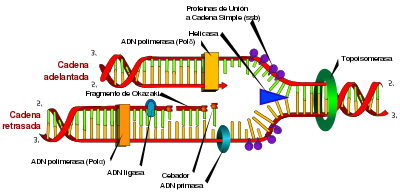
Enzymes called DNA ligases can assemble severed or broken DNA strands. Ligases are particularly important in replication of the discontinuously replicated strand in DNA, as they join together the short DNA fragments generated at the hairpin. replication to form a complete copy of the DNA template. They are also used in DNA repair and in genetic recombination processes.
Topoisomerases and helicases
Topoisomerases are enzymes that have both nuclease and ligase activity. These proteins vary the amount of supercoiled DNA. Some of these enzymes work by cutting the DNA helix and allowing a section to rotate, thus reducing the degree of supercoiling. Once this is done, the enzyme rejoins the DNA fragments. Other types of enzymes are capable of cutting one DNA helix and then passing the second DNA strand through the break, before rejoining the helices. topoisomerases are required for many processes involving DNA, such as DNA replication and transcription.
Helicases are proteins that belong to the group of molecular motors. They use chemical energy stored in nucleoside triphosphates, primarily ATP, to break hydrogen bonds between bases and separate the DNA double helix into single strands. These enzymes are essential for most processes in which enzymes need to access the DNA bases.
Polymerases
Polymerases are enzymes that synthesize nucleotide chains from nucleoside triphosphates. The sequence of its products are copies of existing polynucleotide chains, which are called templates. These enzymes work by adding nucleotides to the 3' hydroxyl group. of the previous nucleotide in a DNA strand. Consequently, all polymerases function in the 5′ → 3′ direction. At the active sites of these enzymes, the nucleoside triphosphate that is incorporated base-pairs with the corresponding one in the template: this allows the polymerase to accurately synthesize the polymerase. strand complementary to the template.
Polymerases are classified according to the type of template they use:
- In DNA replication, DNA-dependent polymerase DNA makes a copy of DNA from a DNA sequence. Precision is vital in this process, so many of these polymerates have a reading verification activity (proofreading). Through this activity, the polymerase recognizes occasional errors in the synthesis reaction, due to the lack of mating between the wrong nucleotide and the mould, which generates a decoupling (Sametch.). If decoupling is detected, an exonucleous activity is activated in the direction 3′ → 5′ and the wrong base is removed. In most organisms, polymerase DNA works in a large complex called replisoma, which contains multiple accessory units, such as helicase.
- The RNA-dependent polymerase DNA are a specialized class of polymerates that copy the sequence of a DNA RNA strand. They include reverse transcribase, which is a viral enzyme involved in retrovirus cell infection, and telomerase, which is necessary for the replication of telomers. Telomerase is an unusual polymerase, because it contains its own RNA mold as part of its structure.
- The transcription is carried out by a DNA-dependent polymerase RNA that copies the sequence of one of the DNA strands in RNA. To start transcribing a gene, polymerase RNA joins a DNA sequence called promoterAnd separate the strands from DNA. Then copy the gene sequence into a transcribed RNA messenger until it reaches a DNA region called finishedwhere it stops and separates from the DNA. As with human-dependent polymerase DNA-dependent polymerase RNA, polymerase II (the enzyme that transcribes most human genomes) functions as a large multiprotein complex that contains multiple regulatory and accessory subunits.
Genetic recombination
 |
 |
A DNA helix does not normally interact with other segments of DNA, and in human cells the different chromosomes even occupy separate areas in the cell nucleus called "chromosomal territories". The physical separation of the different chromosomes is important so that DNA maintains its ability to function as a stable store of information. One of the few times when chromosomes interact is during chromosomal crossover, during which they recombine. Chromosomal crossing over occurs when two DNA strands break, swap, and rejoin.
Recombination allows chromosomes to exchange genetic information and produces new combinations of genes, which increases the efficiency of natural selection and may be important in the rapid evolution of new proteins. During prophase I of meiosis, a Once the homologous chromosomes are perfectly paired, forming structures called bivalents, the phenomenon of crossover or crossover (crossing-over) occurs, in which the non-sister homologous chromatids (from the father and mother) exchange genetic material. The resulting genetic recombination greatly increases the genetic variation between the offspring of sexually reproducing parents. Genetic recombination may also be involved in DNA repair, particularly in the cellular response to double-strand breaks.
The most frequent form of chromosomal crossing over is homologous recombination, in which the two chromosomes involved share very similar sequences. Non-homologous recombination can be harmful to cells, as it can lead to chromosome translocations and genetic abnormalities. The recombination reaction is catalyzed by enzymes known as recombinases, such as RAD51. The first step in the recombination process is a double-strand break, caused either by an endonuclease or by DNA damage. Subsequently, a series of steps catalyzed in part by recombinase, lead to the junction of the two helices forming at least a Holliday junction, in which a segment of a single strand is annealed to the complementary strand in the other helix.. The Holliday junction is a tetrahedral junction structure that can move along the pair of chromosomes, exchanging one strand for another. The recombination reaction is stopped by the cleavage of the junction and the reunion of the released DNA segments.
Evolution of DNA metabolism
DNA contains the genetic information that allows most living things to function, grow, and reproduce. However, it is not clear how long it has served this function in the ~3 billion year history of life, as it has been proposed that the earliest forms of life could have used RNA as genetic material. RNA could have functioned as the central part of a primitive metabolism, since it can transmit genetic information and simultaneously act as a catalyst forming part of ribozymes. This ancient World of RNA where nucleic acids would function as catalysts and how stores of genetic information could have influenced the evolution of the current genetic code, based on four nucleotides. This would be because the number of unique bases in an organism is a compromise between a small number of bases (which would increase the precision of replication) and a large number of bases (which would increase the catalytic efficiency of ribozymes).).
Unfortunately, there is no direct evidence of ancestral genetic systems because the recovery of DNA from most fossils is impossible. This is because DNA is capable of surviving in the environment for less than a million years, and then begins to slowly break down into smaller fragments in solution. Some research claims that older DNA has been obtained, e.g. example a report on the isolation of a viable bacterium from a 250-million-year-old salt crystal, but these data are controversial.
However, tools of molecular evolution can be used to infer the genomes of ancestral organisms from contemporary organisms. In many cases, these inferences are reliable enough that a biomolecule encoded in an ancestral genome can be resurrected in the laboratory for study today. Once the ancestral biomolecule has been resurrected, its properties may offer inferences about primordial environments and lifestyles. This process is related to the emerging field of experimental paleogenetics.
However, the process of working backward from the present has inherent limitations, which is why other researchers try to elucidate the evolutionary mechanism by working from the origin of the Earth onwards. Given enough information about the chemistry in the cosmos, the way in which cosmic substances might have been deposited on Earth, and the transformations that might have taken place on the early Earth's surface, we might be able to learn about the origins for develop models of further evolution of genetic information (see also article on the origin of life).
Common Techniques
Knowledge of the structure of DNA has enabled the development of a multitude of technological tools that exploit its physicochemical properties to analyze its involvement in specific problems: for example, from phylogenetic analysis to detect similarities between different taxa, to the characterization of the individual variability of a patient in their response to a certain drug, going through a global approach, at the genomic level, of any specific characteristic in a group of individuals of interest.
We can classify DNA analysis methodologies into those that seek its multiplication, either in vivo, such as the polymerase chain reaction (PCR), or in vitro >, such as cloning, and those that exploit the specific properties of specific elements, or of properly cloned genomes. This is the case of DNA sequencing and hybridization with specific probes (Southern blot and DNA chips).
Recombinant DNA technology
Recombinant DNA technology, the cornerstone of genetic engineering, makes it possible to propagate large amounts of a piece of DNA of interest, which is said to have been cloned. To do this, said fragment must be introduced into another DNA element, generally a plasmid, which has in its sequence the elements necessary for the cellular machinery of a host, normally Escherichia coli, to replicate it. In this way, once the bacterial strain has been transformed, the cloned DNA fragment reproduces each time it divides.
To clone the DNA sequence of interest, enzymes are used as tools for splicing the fragment and the vector (the plasmid). Said enzymes correspond to two groups: first, restriction enzymes, which have the ability to recognize and cut specific sequences; second, DNA ligase, which establishes a covalent bond between compatible DNA ends (see section Nucleases and ligases).
Sequencing
DNA sequencing consists of elucidating the order of the nucleotides of a DNA polymer of any length, although it is usually directed towards the determination of complete genomes, due to the fact that current techniques allow this sequencing to be carried out at high speed, which which has been of great importance for large-scale sequencing projects such as the Human Genome Project. Other related projects, sometimes the result of collaboration between scientists on a global scale, have established the complete DNA sequence of many animal, plant and microorganism genomes.
The Sanger sequencing method has been the most widely used during the XX century. It is based on the synthesis of DNA in the presence of dideoxynucleosides, compounds that, unlike normal deoxynucleosides (dNTPs), lack a hydroxyl group at their 3' end. Although triphosphated dideoxynucleotides (ddNTPs) can be incorporated into the chain being synthesized, the lack of a 3'-OH end makes it impossible to generate a new phosphodiester bond with the next nucleoside; therefore, they cause the termination of the synthesis. For this reason, the sequencing method is also called "chain termination." The reaction is usually performed by preparing a tube with the template DNA, the polymerase, a primer, standard dNTPs, and a small amount of fluorescently labeled ddNTPs in their nitrogenous base. Thus, ddTTP can be marked in blue, ddATP in red, etc. During polymerization, the growing chains are truncated, randomly, at different positions. Therefore, a series of products of different sizes are produced, the position of the termination coinciding due to the incorporation of the corresponding ddNTP. Once the reaction is finished, it is possible to run the mixture in a capillary electrophoresis (which resolves all the fragments according to their length) in which the fluorescence is read for each position of the polymer. In our example, the reading blue-red-blue-blue would translate as TATT.
Polymerase Chain Reaction (PCR)
Polymerase chain reaction, commonly known as PCR for its acronym in English, is a molecular biology technique described in 1986 by Kary Mullis, whose objective is to obtain a large number of copies of a given DNA fragment, starting from a small amount of it. To do this, a thermostable DNA polymerase is used that, in the presence of a mixture of the four deoxynucleotides, a buffer with the appropriate ionic strength and the cations necessary for the activity of the enzyme, two oligonucleotides (called primers) complementary to the sequence (located at a sufficient distance and in an antiparallel direction) and under suitable temperature conditions, modulated by a device called a thermocycler, exponentially generates new DNA fragments similar to the original and delimited by the two primers.
PCR can be performed as an end-point technique, that is, as a tool for generating the desired DNA, or as a continuous method, in which said polymerization is evaluated in real time. This last variant is common in quantitative PCR.
Southern blot
The «Southern hybridization» method or Southern blot (the original name in the English language) allows the detection of a DNA sequence in a complex or non-complex sample of nucleic acid. To do this, it combines a separation by mass and charge (carried out by gel electrophoresis) with hybridization with a nucleic acid probe labeled in some way (either with radioactivity or with a chemical compound) that, after several reactions, gives rise to to the appearance of a color or fluorescence signal. Said hybridization is carried out after the transfer of the DNA separated by electrophoresis to a filter membrane. A similar technique, but in which the aforementioned electrophoretic separation does not occur, is called dot blot.
The method is named after its inventor, the English biologist Edwin Southern. By analogy to the Southern method, similar techniques have been developed that allow the detection of given RNA sequences (Northern method, which uses RNA probes or labeled DNA) or specific proteins (Western technique, based on the use of antibodies).
DNA chips
DNA chips are collections of complementary DNA oligonucleotides arranged in rows fixed on a support, often made of glass. They are used to study known gene mutations or to monitor gene expression from an RNA preparation.
Applications
Genetic engineering
DNA research has a significant impact, especially in the field of medicine, but also in agriculture and livestock (where the objectives are the same as with the traditional techniques that man has used for millennia —domestication, selection and directed crossings—to obtain more productive animal and plant varieties). Modern biology and biochemistry make intensive use of recombinant DNA technology, introducing genes of interest into organisms, with the aim of expressing a specific recombinant protein, which can be:
- isolated for subsequent use: for example, micro-organisms can be converted into authentic factories that produce large quantities of useful substances, such as insulin or vaccines, which are subsequently isolated and used therapeutically.
- necessary to replace the expression of a damaged endogenous gene that has led to a pathology, which would allow the restoration of the activity of the lost protein and eventually the recovery of the normal, non-pathological physiological state. This is the goal of gene therapy, one of the fields in which medicine is actively working, analyzing advantages and disadvantages of different gene administration systems (viral and non-viral) and the mechanisms of selecting the point of integration of genetic elements (distinct for viruses and transposons) in the target genome. In this case, before considering the possibility of gene therapy in a given pathology, it is essential to understand the impact of the gene of interest in the development of such pathology, for which it is necessary to develop an animal model, eliminating or modifying that gene in a laboratory animal, through the technique knockout. Only in the event that the results in the animal model are satisfactory would the possibility of restoring the damaged gene through gene therapy be analysed.
- used to enrich a food: for example, milk composition (an important source of proteins for human and animal consumption) can be modified by transgenesis, adding exogenous genes and deactivating endogenous genes to improve their nutritional value, reducing infections in the breast glands, providing consumers with antipatogenic proteins and preparing recombinant proteins for their pharmaceutical use.
- useful to improve the resistance of the transformed organism: for example in plants genes that confer resistance to pathogens (virus, insects, fungi...), as well as to abiotic stressors (sillinity, dryness, heavy metals...).
Forensic Medicine
Coroners can use DNA present in blood, semen, skin, saliva, or hair at a crime scene to identify the perpetrator. This technique is called genetic fingerprinting, or also "DNA profiling". In genetic fingerprinting, the length of highly variable sections of repetitive DNA, such as microsatellites, is compared between different people. This method is often very reliable in identifying a criminal. However, identification can be complicated if the scene is contaminated with DNA from different people. The genetic fingerprinting technique was developed in 1984 by the British geneticist sir Alec Jeffreys, and was first used in forensics to convict Colin Pitchfork of the 1983 Narborough and 1986 Enderby murders. Persons accused of certain types of crimes may be required to provide a DNA sample to enter into a database. This has facilitated the work of investigators in resolving old cases, where only a DNA sample was obtained from the crime scene, in some cases allowing a convicted person to be exonerated. Genetic fingerprinting can also be used to identify victims of mass accidents, or to perform consanguinity tests (paternity testing).
Bioinformatics
Bioinformatics involves the manipulation, search, and extraction of information from DNA sequence data. The development of techniques for storing and searching DNA sequences has led to advances in the development of computer software, for many applications, especially phrase search algorithms, machine learning, and database theories. Phrase search or Matching algorithms, which look for the occurrence of a sequence of letters within a larger sequence of letters, were developed to search for specific sequences of nucleotides. In other applications such as text editors, even simple algorithms can work, but DNA sequences can cause these algorithms to exhibit near-worst-case behavior, due to the low number of characters. The related problem of sequence alignment seeks to identify homologous sequences and locate specific mutations that differentiate them. These techniques, primarily multiple sequence alignment, are used when studying phylogenetic relationships and protein function. Collections of data representing genome-sized DNA sequences, such as those produced by the Human Genome Project, they are difficult to use without annotations, which mark the location of genes and regulatory elements on each chromosome. Regions of DNA that have patterns associated with protein- or RNA-coding genes can be identified by gene localization algorithms, allowing researchers to predict the presence of specific gene products in an organism even before it has been experimentally isolated..
DNA nanotechnology

DNA nanotechnology uses the unique molecular recognition properties of DNA and other nucleic acids to create self-assembling branched complexes with useful properties. In this case, DNA is used as a structural material, rather than as a carrier of biological information. This has led to the creation of two-dimensional periodic sheets (both based on tiles, as well as using the method). origami DNA), as well as three-dimensional structures in the shape of polyhedrons.
History, anthropology and paleontology
Over time, DNA stores mutations that are inherited and therefore contains historical information, so by comparing DNA sequences, geneticists can infer the evolutionary history of organisms, their phylogeny. The research Phylogenetics is a fundamental tool in evolutionary biology. By comparing DNA sequences within a species, population geneticists can learn the history of particular populations. This can be used in a wide variety of studies, from ecology to anthropology, as illustrated by the DNA analysis carried out to identify the Ten Lost Tribes of Israel. On the other hand, DNA is also used to study recent family relationships..
Similarly in paleontology (in paleogenetics) DNA in some cases can also be used to study extinct species (fossil DNA).
Contenido relacionado
Spermatophyte
Tragopogon pratensis
Van der Waals radius

















![Animación de la estructura de una sección de ADN. Las bases se encuentran horizontalmente entre las dos hebras en espiral. Versión ampliada[52]](https://upload.wikimedia.org/wikipedia/commons/1/16/DNA_orbit_animated.gif)