Ribonucleic acid
Ribonucleic acid (RNA) is a nucleic acid made up of a chain of ribonucleotides. It is present in both prokaryotic and eukaryotic cells, and is the unique genetic material of certain viruses (RNA viruses).
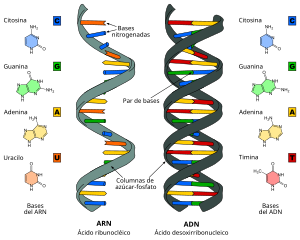
RNA can be defined as a molecule made up of a single chain of ribonucleotides, each one made up of ribose, a phosphate, and one of the four nitrogenous bases (adenine, guanine, cytosine, and uracil). Cellular RNA is linear and single-stranded (single-stranded), but in the genome of some viruses it is double-stranded.
In cellular organisms it performs various functions. It is the molecule that directs the intermediate stages of protein synthesis; DNA cannot act alone, and relies on RNA to transfer this vital information during protein synthesis (production of proteins needed by the cell for its activities and development). Various types of RNA regulate gene expression, while that others have catalytic activity. RNA is thus much more versatile than DNA.
Discovery and history
Nucleic acids were discovered in 1867 by Friedrich Miescher, who named them nuclein, since he isolated them from the cell nucleus. Later, it was found that prokaryotic cells, which lack a nucleus, also contained nucleic acids. The role of RNA in protein synthesis was suspected in 1939. Severo Ochoa won the 1959 Nobel Prize in Medicine after discovering how RNA was synthesized.
In 1965 Robert W. Holley found the 77-nucleotide sequence of a yeast transfer RNA, for which he was awarded the Nobel Prize in Medicine in 1968. In 1967, Carl Woese verified the catalytic properties of some RNAs and suggested that early life forms used RNA as a carrier of genetic information as well as a catalyst for their metabolic reactions (RNA world hypothesis). In 1976, Walter Fiers and his collaborators determined the complete RNA sequence of the genome of a RNA virus (MS2 bacteriophage).
In 1990 it was discovered in Petunia that introduced genes can silence similar genes in the same plant, leading to the discovery of RNA interference. At about the same time, micro-RNAs were found, small 22-nucleotide molecules that played a role in the development of Caenorhabditis elegans. The discovery of RNAs that regulate gene expression has allowed the development of drugs made from RNA, such as small interfering RNAs that they silence genes.
In the year 2016 it is practically proven that RNA molecules were the first form of life itself to inhabit planet Earth (RNA world hypothesis).
RNA biochemistry
Like DNA, RNA is made up of a chain of repeating monomers called nucleotides. Nucleotides are linked one after the other by negatively charged phosphodiester bonds.
Each nucleotide is made up of three components:
- A five-carbon monosaccharide (pentosa) called β-D-ribofuranosa.
- A phosphate group
- A nitrogen base, which can be
- Adenine (A)
- Cytosine (C)
- Guanina (G)
- Uracilo (U)
| RNA | DNA | |
|---|---|---|
| Pentosa | Ribosa | Desoxirribosa |
| Pure | Adenina and Guanina | Adenina and Guanina |
| Pirimidins | Cytosine and Uracilo | Cytosine and Timina |
The carbons of ribose are numbered from 1' at 5' Clockwise. The nitrogenous base is attached to carbon 1'; the phosphate group is attached to the 5' and carbon 3' of the ribose of the next nucleotide. The peak has a negative charge at physiological pH, which gives the RNA a polyanionic character. Purine bases (adenine and guanine) can form hydrogen bonds with pyrimidine bases (uracil and cytosine) according to the C=G and A=U scheme. In addition, other interactions are possible, such as base stacking or paired tetraloops G=A. Many RNAs contain, in addition to the usual nucleotides, modified nucleotides, which originate by transformation of the typical nucleotides; they are characteristic of transfer RNA (tRNA) and ribosomal RNA (rRNA); methylated nucleotides are also found in eukaryotic messenger RNA.
Double mating
The hydrogen bonding interaction described by Watson and Crick forms base pairs between a purine and a pyrimidine. This pattern is known as Watson and Crick mating. In it, adenine pairs with uracil (thymine, in DNA) and cytosine with guanine. However, many other forms of mating occur in RNA, of which the most ubiquitous is wobble mating (also wobble mating or hesitant mating) for the pair G-U. This was first proposed by Crick to explain codon-anticodon pairing in tRNAs and has been confirmed in almost all RNA classes in all three phylogenetic domains.
Structure
Primary structure
Refers to the linear sequence of nucleotides in the RNA molecule. The following structural levels (secondary, tertiary structure) are a consequence of the primary structure. Furthermore, the sequence itself may be functional information; This can be translated to synthesize proteins (in the case of mRNA) or function as a recognition region, catalytic region, among others.
Primary structure of tRNAPhe
Secondary structure
RNA folds as a result of the presence of short intramolecular base-pairing regions, that is, base pairs formed by more or less distant complementary sequences within the same strand. Secondary structure refers, then, to base-pairing relationships: “The term 'secondary structure' denotes any flat pattern of base-pairing contacts. It is a topological concept and should not be confused with some kind of two-dimensional structure". Secondary structure can be described from structural reasons which are usually classified as follows:
Tertiary Structure
The tertiary structure is the result of the interactions in space between the atoms that make up the molecule. Some interactions of this type include base stacking and base pairings other than those proposed by Watson and Crick, such as the Hoogsteen pairing, triple pairings, and ribose zippers.
Helix A
Unlike DNA, RNA molecules are usually single-stranded and do not form extensive double helices, however, in base-paired regions it does form helices as a tertiary structural motif. An important structural feature of RNA that distinguishes it from DNA is the presence of a hydroxyl group at the 2' position. of ribose, which causes RNA double helices to adopt an A conformation, rather than the B conformation that is more common in DNA. This A helix has a very deep major groove and narrow and a wide and shallow minor groove. A second consequence of the presence of this hydroxyl is that the phosphodiester bonds of RNA in regions where double helixes do not form are more susceptible to chemical hydrolysis than those of DNA; RNA phosphodiester bonds hydrolyze rapidly in alkaline solution, whereas DNA bonds are stable. The half-life of RNA molecules is much shorter than that of DNA, a few minutes in some bacterial RNAs or a few days. in human tRNAs.
Biosynthesis
RNA biosynthesis is normally catalyzed by the enzyme RNA polymerase using a strand of DNA as a template, a process known as transcription. Therefore, all cellular RNAs come from copies of genes present in the DNA.
Transcription begins with the recognition by the enzyme of a promoter, a characteristic sequence of nucleotides in the DNA located before the segment to be transcribed; the DNA double helix is opened by the helicase activity of the enzyme itself. The RNA polymerase then progresses along the DNA strand in the 3' sense. → 5', synthesizing a complementary RNA molecule; this process is known as elongation, and the growth of the RNA molecule occurs in the 5' direction. → 3'. The DNA nucleotide sequence also determines where RNA synthesis ends, thanks to the fact that it has characteristic sequences that RNA polymerase recognizes as termination signals.
After transcription, most RNAs are modified by enzymes. For example, a modified guanine nucleotide (7-Methylguanosine) is added to the newly transcribed eukaryotic pre-messenger RNA at the 5' end. via a triphosphate bridge forming a 5'→ 5' unique, also known as "hood" or "hood", and a long sequence of adenine nucleotides at the 3' end (poly-A tail); later the introns (non-coding segments) are removed in a process known as splicing or splicing.
In viruses, there are also several RNA-dependent RNA polymerases that use RNA as a template for the synthesis of new RNA molecules. For example, several RNA viruses, such as polioviruses, use this type of enzyme to replicate their genome.
RNA classes
Messenger RNA (mRNA) is the type of RNA that carries information from DNA to ribosomes, the site of protein synthesis. The nucleotide sequence of the mRNA determines the amino acid sequence of the protein. Therefore, the mRNA is called the coding RNA.
However, many RNAs do not code for proteins, and are called non-coding RNAs; They originate from self genes (RNA genes), or are the introns rejected during the splicing process. Noncoding RNAs include transfer RNA (tRNA) and ribosomal RNA (rRNA), which are key elements in the translation process, and various types of regulatory RNAs.
Certain non-coding RNAs, called ribozymes, are capable of catalyzing chemical reactions such as cutting and splicing other RNA molecules, or forming peptide bonds between amino acids in the ribosome during protein synthesis.
RNA involved in protein synthesis
Messenger RNA
Messenger RNA (mRNA) is what carries information about the protein's amino acid sequence from the DNA, where it is written, to the ribosome, where the cell's proteins are synthesized. It is therefore an intermediary molecule between DNA and protein, and the name "messenger" is entirely descriptive. In eukaryotes, mRNA is synthesized in the nucleoplasm of the cell nucleus and where it is processed before accessing the cytosol, where ribosomes are found, through the pores of the nuclear envelope.
RNA transfer
Transfer RNAs (tRNAs) are short polymers of about 80 nucleotides, which transfer a specific amino acid to the growing polypeptide; bind to specific sites on the ribosome during translation. They have a specific site for amino acid attachment (3' end) and an anticodon made up of a nucleotide triplet that hydrogen bonds to the complementary codon of the mRNA. These tRNAs, like other types of RNA, they can be post-transcriptionally modified by enzymes. The modification of any of its bases is crucial for the decoding of mRNA and for maintaining the three-dimensional structure of the tRNA.
Ribosomal RNA
Ribosomal RNA (rRNA) is combined with proteins to form ribosomes, where it represents about 2/3 of the ribosomes. In prokaryotes, the larger subunit of the ribosome contains two rRNA molecules and the smaller subunit contains one. In eukaryotes, the largest subunit contains three rRNA molecules and the smallest one. In both cases, specific proteins are associated on the framework constituted by the mRNAs. rRNA is very abundant, accounting for 80% of the RNA found in the cytoplasm of eukaryotic cells. Ribosomal RNAs are the catalytic component of ribosomes; They are responsible for creating the peptide bonds between the amino acids of the polypeptide being formed during protein synthesis; They therefore act as ribozymes.
Regulatory RNAs
Many types of RNA regulate gene expression by being complementary to specific regions of mRNA or DNA genes.
RNA interference
RNA interference (RNAi) are RNA molecules that suppress the expression of specific genes through mechanisms known globally as ribointerference or RNA interference. Interfering RNAs are small molecules (20 to 25 nucleotides) that are generated by fragmentation of longer precursors. They can be classified into three large groups:
Micro-RNA
Micro-RNAs (miRNAs) are short chains of 21 or 22 nucleotides found in eukaryotic cells that are generated from specific precursors encoded in the genome. Upon transcription, they fold into intramolecular hairpins and then bind to enzymes forming an effector complex that can block mRNA translation or accelerate its degradation beginning with enzymatic removal of the poly A tail.
Small interfering RNA
Small interfering RNAs (siRNA or siRNA), made up of 20-25 nucleotides, are frequently produced by viral RNA cleavage, but may also be of endogenous origin. After transcription, they assemble into a protein complex called RISC (RNA-induced silencing complex) that identifies complementary mRNA that is cut into two halves that are degraded by cellular machinery, thus blocking gene expression.
Piwi-associated RNA
The RNAs associated with Piwi are chains of 29-30 nucleotides, typical of animals; they are generated from long single-stranded (consisting of a single strand) precursors, in a process that is independent of Drosha and Dicer. These small RNAs associate with a subfamily of the "Argonaut" called Piwi proteins. Germ line cells are active; they are thought to be a defensive system against transposons and to play some role in gametogenesis.
Antisense RNA
An antisense RNA is the complementary (non-coding) strand of an mRNA (coding) strand. Most inhibit genes, but a few activate transcription. Antisense RNA pairs with its complementary mRNA to form a double-stranded molecule that cannot be translated and is enzymatically degraded. Introduction of a transgene encoding antisense mRNA is a technique used to block the expression of a gene of interest. Radioactively labeled antisense mRNA can be used to show the level of gene transcription in various cell types. Some antisense structural types are experimental, as they are used as antisense therapy.
Long noncoding RNA
Many long non-coding RNAs (long ncRNAs) regulate gene expression in eukaryotes; one of these is Xist, which covers one of the two X chromosomes in female mammals, inactivating it (Barr's corpuscle). Various studies reveal that it is active at low levels. In certain cell populations, a quarter of the protein-coding genes and 80% of the lncRNAs detected in the human genome are present in one or no copies per cell, as there is a restriction on certain RNAs.
Riboswitch
A riboswitch is a part of mRNA (messenger ribonucleic acid) to which small molecules can bind that affect gene activity. An mRNA containing a riboswitch is therefore directly involved in regulating its own activity which depends on the presence or absence of the signaling molecule. Such riboswitches are found in the 5' (5'-UTR), located before the start codon (AUG), and/or in the 3' (3'-UTR), also called a drag sequence, located between the termination codon (UAG, UAA or UGA) and the poly A tail.
RNA with catalytic activity
Ribozymes
RNA can act as a biocatalyst. Certain RNA associates with proteins forming ribonucleoproteins and it has been verified that it is the RNA subunit that carries out the catalytic reactions; these RNAs carry out the reactions in vitro in the absence of protein. Five types of ribozymes are known; three of them carry out self-modifying reactions, such as intron deletion or self-cleavage, while the others (ribonuclease P and ribosomal RNA) act on different substrates. Thus, ribonuclease P cuts a precursor RNA into tRNA molecules, while that ribosomal RNA performs the peptide bond during ribosomal protein synthesis.
Spliceosome
Introns are separated from pre-mRNA during a process known as splicing by spliceosomes, which contain numerous small nuclear RNAs (snRNAs). In other cases, the introns themselves act as ribozymes and they separate themselves from the exons.
Nucleolar small RNA
Small nucleolar RNAs (sRNAs), found in the nucleolus and in the bodies of Cajal, direct the modification of nucleotides of other RNAs; the process consists of transforming one of the four typical nitrogenous bases (A, C, U, G) in others. pRNAs associate with enzymes and guide them by pairing with specific sequences of the RNA they will modify. rRNA and tRNA contain many modified nucleotides.
Mitochondrial RNA
Mitochondria have their own protein synthesis apparatus, which includes rRNA (in ribosomes), tRNA, and mRNA. Mitochondrial RNAs (mtRNA or mtRNA) represent 4% of total cellular RNA. They are transcribed by a specific mitochondrial RNA polymerase.
RNA genomes
DNA is the molecule that carries genetic information in all cellular organisms, but, like DNA, RNA can store genetic information. RNA viruses completely lack DNA and their genome is made up of RNA, which encodes the virus's proteins, such as those of the capsid, and some enzymes. Said enzymes carry out the replication of the viral genome. Viroids are another class of pathogens that consist exclusively of an RNA molecule that does not code for any protein and that is replicated by the host cell's machinery.
RNA world hypothesis
The RNA world hypothesis proposes that RNA was the first nucleic acid to appear on Earth, later preceding DNA, and in turn these nucleic acids together with proteins, when united with spontaneously formed liposomes, would give rise to the first cells. It is based on the verification that RNA can contain genetic information, in a similar way to DNA, and that some types are capable of carrying out metabolic reactions, such as self-cleavage or peptide bond formation.
For years there has been speculation about which came first, DNA or enzymes, since enzymes are synthesized from DNA and DNA synthesis is carried out by enzymes. If it is assumed that the first forms of life used RNA both to store their genetic information and carry out their metabolism, this obstacle is overcome. Experiments with basic ribozymes, such as Q-beta viral RNA, have shown that simple self-replicating RNA structures can resist even strong selective pressures (such as chain terminators of opposite chirality).
Contenido relacionado
Trace element
Zygophyllaceae
Onagraceae