Polymerase chain reaction
The polymerase chain reaction technique (in English, polymerase chain reaction or PCR) is a molecular biology technique developed in 1986 by Kary Mullis. Its goal is to obtain a large number of copies of a particular DNA fragment, starting from a minimum; in theory it is enough to start with a single copy of that original fragment, or mold.
The technique, for a better understanding of the text, can be explained in the following steps in a simple way:
- Combine a single synthetic strand of only 20 bases.
- Separate DNA strands to investigate and see if the synthetic strand fits somewhere in these.
- It does: You can "animate" the synthetic small strand to be reproduced, providing some more bases, and the appropriate enzymes and conditions to do the task.
- If a sufficient number of times is made, this fragment will grow from a microscopic scale to a macroscopic scale.
- Then another synthetic cut is produced with a sequence of different bases and only the process is repeated.
- In the end you can read the entire genome.
With this procedure, it is much easier to identify, with a very high probability, viruses or bacteria that cause a disease, identify people (corpses) or do scientific research on amplified DNA. These uses derived from amplification have made it a very widespread technique, both in the forensic field and in chemical laboratories, with the consequent lower cost of the equipment necessary to carry out said technique.
Base and importance
This technique is based on the natural property of DNA polymerases to replicate DNA strands, for which cycles of alternate high and low temperatures are used to separate the newly formed DNA strands from each other after each replication phase and, then let the DNA strands come back together so they can be duplicated again. The polymerase chain reaction was perfected by Kary Mullis of the Cetus Corporation in California in the 1980s. Initially, the technique was slow, since the polymerases denatured when making temperature changes and it was necessary to add new polymerases in each cycle. Since the temperatures of the cycle (95 °C in the DNA denaturation phases) imply the immediate denaturation of all proteins, thermostable DNA polymerases are used, extracted from microorganisms adapted to living at these temperatures, which are restrictive for most living beings.. These microorganisms, generally archaea, are: Thermus aquaticus (Taq polymerase), Pyrococcus furiosus (Pfu), Thermococcus litoralis (Vent) and Thermus thermophilus (Tth). Mixtures of very processive polymerases (Taq) are generally used with others capable of correcting errors (Pfu, Vent).
Today, the entire PCR process is automated by means of a device called a thermocycler, which allows heating and cooling the reaction tubes to control the temperature necessary for each stage of the reaction. Many modern thermal cyclers make use of the Peltier effect, which allows both heating and cooling tubes simply by reversing the electrical current. The tubes used for PCR have a very thin wall, which favors good thermal conductivity, allowing thermal equilibrium to be reached quickly. Almost all thermal cyclers have a system that heats the closure lid in order to avoid condensation on the reaction tubes. Older thermal cyclers lacked this system and solved the problem of condensation with a layer of oil on top of the reaction mix or with a little wax inside the tubes. There are currently some thermal cyclers that use or can use mineral oil in the PCR tube such as the new generation airflow thermal cyclers.
PCR is generally a common and often indispensable technique in medical and biological research laboratories for a wide variety of applications. These include DNA cloning for sequencing, DNA-based phylogeny, gene functional analysis, diagnosis of inherited disorders, fingerprinting (used in forensics and paternity testing), and detection and diagnosis. of infectious diseases.
Reagents
To perform the technique you will need:
- The 4 desoxyribonucleotides-triphosphate (dNTP), substrates to polymerize new DNA.
- Two pumps or initiators, oligonucleotides that are, each, complementary to one of the two strands of DNA. They are short sequences, between six and forty nucleotides, usually from eighteen to twenty-two, which allow the polymerase to initiate the reaction. They must be confronted and not far away. They delimit the DNA zone to amplify, that is to say, they correspond to the nucleotides that define the ends of the sequence that you want to replicate.
- Divalent ions. Magnesium is usually used (Mg2+), commonly added as magnesium chloride (MgCl2), or some other divalent cathion. You can also use manganese (Mn2+), for DNA mutagenesis using PCR, since high Mn concentrations2+ increase the error rate during DNA synthesis. They act as cofactors of the polymerase.
- Monovalent ions, like potassium.
- A tampon solution or buffer that keeps the pH suitable for the operation of the polymerase DNA.
- Polymerase DNA or mixture of different polymerase with optimal temperature around 70 °C (the most common is Taq polymerase).
- DNA mould, which contains the DNA region to be amplified.
- Thermocoupler, the device that maintains the necessary temperature in each of the stages that make up a cycle.
PCR concepts
- Sensitivity: refers to the minimum amount of DNA required for the amplification, that is, to get a band. It is related to false negatives, since a sample may be positive but given as negative because it has not been amplified because it does not have enough DNA.
- Specificity: refers to obtaining a single amplified product. It is determined by the oligos and the specificity with which they bind to the mould DNA. This way, if the oligos have more than one place to which they can be joined will appear more than one amplified product. It relates to false positives, since a sample may be negative but given as positive because a non-danger DNA region has been amplified or not intended to be amplified.
- Efficiency: refers to the maximum amplification that can be obtained in a certain number of cycles.
- Fidelity: refers to the mistakes made by polymerase DNA during amplification. This concept is of particular importance in sequencing, but in other cases it is not so important. Good fidelity allows you to avoid false positives and/or negatives.
Amplification Cycle
The PCR process typically consists of a series of 20 to 35 repeated temperature changes called cycles; each cycle usually consists of 2-3 steps at different temperatures. Common PCR is performed with cycles that have three temperature steps. Cycling steps are often preceded by a thermal shock (called a "hold") at high temperature (>90 °C), and followed by another hold at the end of the process for final product extension or short storage. The temperatures used and the time applied in each cycle depend on a wide variety of parameters. These include the enzyme used for DNA synthesis, the concentration of divalent ions and dNTPs in the reaction, and the junction temperature of the primers, as well as the length of the DNA to be amplified.
Currently, almost all thermal cyclers give the option of performing the PCR reaction with the so-called “hot cap”. That is, the thermocycler system will apply heat to the top of the vial containing the PCR mix. At the beginning, the laboratories that began to use the first devices that were commercialized and that did not include this system had to put a few drops of oil inside the vial. The objective of this procedure, like that of the hot lid, is to avoid condensation of the sample, since there are two phases in the eppendorf: liquid and gas. As the sample condenses, we lose volume of the mixture. However, by heating the lid or adding drops of oil we avoid this physical process, keeping the volume of the sample almost intact.
Home
This step consists of bringing the reaction to a temperature of 94-96 °C (or 98 °C if using an extreme thermostable polymerase), which is maintained for 1-9 minutes. This is only necessary for DNA polymerases that require heat activation.
Denaturation
First, the DNA is denatured (the two strands of which it is made are separated). This step can be done in different ways, heating (94−95 °C) of the sample being the most common way. The temperature at which it is decided to carry out the denaturation depends, for example, on the proportion of G+C that the chain has, as well as its length. Other methods, rarely used in the PCR technique, would be the addition of salts or chemical agents capable of denaturing.
Primer alignment or binding
This is followed by primer hybridization, that is, the primer will bind to its complementary sequence in the template DNA. To do this, it is necessary to lower the temperature to 40−68 °C for 20-40 seconds (depending on the case), thus allowing alignment. Stable hydrogen bonds between DNA strands (DNA-DNA junction) are only formed when the primer sequence is very similar to the template DNA sequence. The polymerase joins the hybrid of the template strand and the primer, and begins to synthesize DNA. The primers will act as boundaries for the region of the molecule to be amplified.
Extension or elongation of the chain
The polymerase acts, taking the template DNA to synthesize the complementary strand and starting from the primer as the initial support necessary for the synthesis of new DNA. The polymerase synthesizes a new DNA strand complementary to the template strand by adding the complementary dNTPs in the 5'→ 3' direction, linking the 5'-phosphate group of the dNTPs with the 3'-hydroxyl group from the end of the growing (extending) DNA strand. The temperature for this step depends on the DNA polymerase we use. For Taq polymerase, the temperature of peak activity is 75−80 °C (commonly 72 °C ). The extension time depends both on the DNA polymerase used and on the length of the DNA fragment to be amplified. There is a commonly used rule of thumb: at its optimum temperature, DNA polymerase will polymerize a thousand bases in one minute.
Final elongation
Single step run at 70−74 °C for 5-15 minutes after the last PCR cycle. It ensures that any remaining single-stranded DNA is fully amplified.
Conservation
This is a step that is performed at 4−15 °C for an indefinite time to preserve the short-term reaction.
PCR is typically performed with a reaction volume of 15−100 μL, in small 0.2−0.5 mL which are placed in the thermocycler.
To verify that the PCR has generated the expected DNA fragment, electrophoresis techniques are used, which separate the generated DNA fragments according to their charge, that is, length, and, to a lesser extent and depending on the matrix used, to its size: typically agarose gel electrophoresis is used, for large fragments; in acrylamide, for the little ones; and, faster and more applicable to PCR associated with fluorescent labeling, capillary electrophoresis. The size(s) of the PCR products are determined by a marker of molecular weight of DNA, which contains DNA fragments of known size, and which is run on the gel together with the PCR products.
PCR optimization
In practice, PCR can fail for a number of reasons, including:
- The RCP is a highly sensitive technique, that is, it needs a minimum amount of DNA to get a large number of copies. In addition, it can be very prone to errors if it is carried out in inadequate sterility conditions, which lead to DNA amplification not corresponding to the sample to analyze (and therefore to uncertain conclusions). A series of precautions should be taken to prevent the pollution with strange DNA. Examples in which it is especially important to avoid amplification are the detection of diseases (not to misdiagnose patients) or the diagnosis of identity and kinship. Possible precautions are:
- Be especially careful during the steps leading to amplification: collection, shipping, custody and processing of samples.
- Exhaustive cleaning and sterilization (lejía, etanol, UV light, psoralenos...) of the work surface between the realization of a PCR and the following.
- In genetic diagnostic laboratories, separate areas of work are usually given: a laboratory for preparing the mother mixture for the RCP, another for adding the DNA to amplify, another where the machine is found to perform the RCP and another where the already amplified sample is opened and analyzed.
- Biosafety pods to reduce vapours loaded with disease amplyons.
- Appropriate protection of operators who manipulate the samples and instruments of the laboratory: gloves, robe, covers, safety glasses, hair collected... In molecular diagnosis, it is desirable that these elements be changed by moving from one working area to another.
- Positive and negative controls.
- Clothing of all the lab staff. In case of finding an unanticipated amplification you can check if it comes from one of the operators.
- The techniques barley design are important in improving PCR products and avoiding the formation of false products. Some considerations when designing these barleys are:
- It is recommended to use initiators of more than 15 nucleotides (18 - 30 nn) Normally, 20 nucleotides are usually designed. Very short barleys make our RCP not very specific, and those that exceed in length will cause us to lose performance in the reaction.
- Pumpers should not differ in more than 3 bases.
- The proportion between the Pyrimidian and Pyrimidian bases of the two oligos is 1:1 (40-60 % at most), and start and end with 1 or 2 polymer bases.
- A homogeneous distribution of the four nucleotides in the sequence is required, avoiding the polyT/A/G/C.
- The Tm (melting temperature) of the oligos cannot differ in more than 5 °C.
- Pumpers should not include in their sequence regions that are complementary to each other, or digits will be formed between them.
- There is a wide variety of Polymerates available, can choose the one that best fits our needs. For example, some have non-existent activities in others or perform them more efficiently, or operate at temperature intervals where others are denaturalized or lose functionality. The most common are taq and pfu.
- Components of reaction tampon must conform to the requirements of our enzymes.
- The thermocoupler, of course, it must be able to correctly reproduce the cycles of the RCP: for this purpose, various commercial houses will offer us thermocouplers elaborated with materials that make development more effective and the course of the stages, in addition to various management facilities. Within the laboratory, its management, maintenance and location will be key.
Apart from aspects such as contamination and some failure in primer annealing, there may be other complexities that affect PCR, such as:
- DNA degradation: This can occur when the sample is manipulated under sub-optimal conditions or when an autopsy is done, for example, and results in the absence of amplifying or partial results. A particular case is the "allele dropout", or loss of an allele, which can be confused with being homozygotic to said allele.
- PCR inhibition: There may be agents who interfere in the correct course of the RCP. You will require additional sample processing to remove these compounds from the medium before preparing the PCR mix. Both blood and semen contain this type of inhibitors, so it is necessary a prior purification.
- DNA modification: Substances such as formol (based on tissue conservation) or ultraviolet light modify the DNA, thus altering the results of the amplification.
- Artefacts: situations like the "stutter" or "shadow" bands can occur, which consist of the polymerase amplifying a repetition of more than one microsatellite or tandem repetition.
- Alelo out of pattern: an amplified allele may not match the pattern of reference peaks. This is a sign that something went wrong during the RCP.
- Mixes: it is possible that two show are mixed in some way and the result is a combination of the beak pattern that can be expected after the amplification of each one of them separately. This and the previous case makes it difficult to interpret the results.
Types of PCR
Nested PCR
Very sensitive PCR technique in which the product of an amplification is used as a template to carry out a second amplification with primers that are located within the first amplified sequence, that is, when we have the first, how its amplification can be unite the primers and an amplification is made again within the initial amplicon. This type of PCR has the advantage of providing high sensitivity and specificity. The specificity increases because as it is amplification of a previously obtained amplicon, the primers will only hybridize in one site within the molecule and the result will be a single band. Thus, we avoid possible unspecific hybridizations of the primers. The disadvantage of this technique is that it does not allow us to quantify the sample.
PCR Overlap Extension (Mutagenesis)
Sequence changes are introduced into (cloned) fragments of DNA. 2 mutagenic and 2 other primers are required. A 5' and a fragment 3' that overlap, both carrying the mutation. The products are used in another reaction to produce the full length mutated DNA.
PCR in situ
PCR in situ consists of a PCR reaction on histological sections or cells, where the generated products can be visualized at the amplification site. It is performed on fixed preparations on a slide. In the in situ PCR technique, a first amplification of target DNA is performed and then detection by means of conventional in situ hybridization with DNA/RNA probes. In this way, very small amounts of genome can be detected. This technology is powerful in the ability to specifically amplify a population of underrepresented sequences.
Multiple PCR
PCR in which more than one sequence is amplified simultaneously. To do this, two or more pairs of primers are combined in the same tube, together with the rest of the reaction reagents in sufficient amounts, to simultaneously amplify several DNA segments. Advantages: information on several loci in a single reaction, less template for analysis, less reagents, fast database construction. Disadvantages: To carry it out properly and without errors, careful optimization of the process is required.
Reverse Transcriptase PCR (RT-PCR)
It is a variant of PCR in which we use RNA as the initial template instead of DNA, and it uses a reverse transcriptase (such as Tth) to carry out the synthesis of a DNA complementary to the RNA (cDNA). Thus, the initial development of an RT-PCR would be:
- 1.♪ step: retrotranscription from RNA.
- 2nd step: amplification from the first strand of DNAc.
- 3.♪ step: Standard PCR.
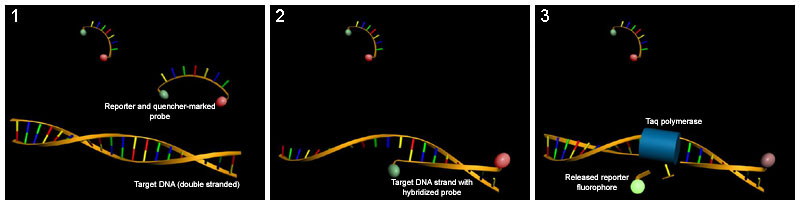
Real-time PCR or quantitative PCR (qPCR)
PCR reaction whose main characteristic is that it allows to quantify the amount of DNA or RNA present in the original sample, or to identify with a very high probability, specific DNA samples based on their melting temperature (also called value < i>Tm, from English melting temperature).
It can be divided into techniques based on non-specific fluorochromes and techniques based on specific probes.
In the techniques based on fluorochromes, the DNA, whose quantity multiplies with each cycle, binds to the fluorochrome (usually SYBR Green) producing fluorescence that is measured by the thermocycler suitable for real-time PCR. It allows quantifying only one sequence per reaction but has the advantage of using normal primers for its implementation. It is much cheaper than the one that uses specific probes.
The techniques based on specific probes use a probe linked to two fluorochromes that anneals in the intermediate region between the forward (forward) and reverse ( reverse); when the probe is intact, it exhibits fluorescence resonance energy transfer (FRET). Said FRET does not occur when the probe is damaged and the two fluorochromes are distant, due to the 5'-3' activity of the probe. DNA polymerase exonuclease. This allows to monitor the change of the fluorescence pattern and to deduce the level of amplification of the gene.
Most of these drawbacks have been solved with the introduction of real-time PCR (Q-PCR), which eliminates any post-PCR process as it monitors the amplification progression as it occurs. Unlike conventional (end-point) PCR, which measures DNA accumulation at the end of a predetermined number of cycles, with Q-PCR this is done during the amplification process using fluorescence, such that its increase is proportional to the amount of DNA formed. The process can be easily automated using a system that performs the amplification (thermocycler) and that in turn is capable of reading fluorescence. There is a wide range of devices on the market. Most of them can work with the various fluorescent labeling options and are "open", that is, they allow the amplification and reading conditions to be programmed so that their use is not limited to certain reagents.
Variations on the basic PCR
- Ice-specific PCR: This diagnostic or cloning technique is used to identify or use single-base polymorphisms (SNPs). It uses specific strainers for normal and mutant sequences. The most common design is an analysis in two tubes with two barleys: one normal and another mutant in separate reactions along with the control pumps.
- PCR "assembly": consists of the artificial synthesis of long DNA sequences, making for it PCR in a long oligonucleotide fund with short overlapping sequences.
- Asymmetric PCR: used to preferably amplify an original DNA string with respect to the other.
- Colony PCR: through this technique, bacterial colonies Escherichia coli can be quickly examined for viable DNA vector constructions.
- Amplification dependent on helicase: This technique is very similar to conventional RCP, but it uses helicase enzyme and constant temperature instead of DNA polymerase and repeated cycles of hybridization-longation.
- PCR hot-start: This technique reduces unspecified amplification during the initial stages of the RCP: while the machine reaches the temperature of the first stage (about 95 °C) it may occur the union of the barbers and there is amplification, since on the way to reach these 95 °C it passes through the temperature of ringing (which is lower).
- To avoid this, the PCR hot-start is based on the fact that the reaction begins when the machine is already at 95°, due to the fact that the polymerase or magnesium chloride are not present, which we can get through various techniques:
- - Add polymerase or magnesium chloride after the warming period
- - Separate by a layer of wax the different components of the reaction. The wax melts when reaching 95 °C and is then when the components come in contact
- - Antipolymerase antibodies that are blocking the polymerase. By reaching 95 °C these antibodies are inactivated because they are denaturalized and polymerase can act
- Specific intersecution PCR (ISSR): This is a PCR method for use in genetic fingerprints, which amplifies regions between simple sequence repetitions to produce a unique genetic footprint of amplified fragment lengths.
- Reverse PCR: is a method used to perform the PCR when only an internal sequence is known. Very useful in identifying sequences that flank genomic inserts.
- RCP mediated by ligation: This method uses small DNA linkers linked to interest DNA and multiple barbers hybridizing these linkers.
- Specific methylation PCR (MSP): is used to detect genetic DNA methylation on CpG islands.
- Multiple expansion Dependant Sonda (Multiplex Ligation-dependent Probe Amplification or MLPA): allows to amplify several target sequences with a single pair of barleys, thus avoiding the resolution limitations of the multiplex PCR.
- Quantitative RCP: is used to measure the amount of a PCR product. In the classic PCR technique, it is quantified by approximation with dilutions and amplification of known concentrations of the target sequence. It can also be quantified by competitive method: it works with growing concentrations known of a fragment that can be amplified by the same oligos as the sample of study, but smaller than this, so that with the results obtained it can be estimated which concentration of the competitor is equivalent to that of the sample of unknown quantity. PCR quantification is optimized in real-time PCR technique.
- PCR-TAIL: The asymmetrical interlacing thermal PCR is used to isolate an unknown sequence that flanks a known sequence.
- PCR touchdown: This is a variant of the RCP that is used when the exact sequence of the ends of the sequence is unknown to amplify, so it is assumed that there can be some uneven base in the barbeter-sequence alignment. Its purpose is to reduce the non-specific background by gradually lowering the hybrid temperature along the progress of the RCP.
- PAN-AC: This method uses isoterm conditions for amplification, and can be used in living cells.
- Fast thermal cycle for PCR:DNA amplification can be performed quickly, such as completing 30 cycles in less than 30 minutes: Rapid thermal cycle. It has normally been considered that the stages of each cycle are three reactions that occur in three separate periods and at three different temperatures. This sequential equilibrium paradigm does not take into account that the sample temperature does not change instantly, in fact, during a RCP the sample is most of the time in transition temperatures. The rapid thermal cycle takes advantage of instant denaturalization and hybridization, whose temperatures need to be reached, but not maintained. In addition, for short products, the extension can be carried out during the transition to the extension temperature, and therefore it is not necessary to keep it. A fast cycle could then be described as 94 °C, 0 s; 55 °C, 0 s and 72 °C, 0 s. As we see, this model does not help, because it takes us only to extreme temperatures and gives us no information about what happens between them. Instead, in the kinetic paradigm for fast-cycle PCR the sample temperature history is completely described. Denaturalization, hybridization and elongation are considered to occur in a temperature range and can also be temporarily overlapped. There are thermocyclers that allow a cycle to be completed in 20-60 seconds, so 30 cycles take 10 to 30 minutes.
Applications
The PCR technique has many applications: already in basic science, as a tool for detection and/or generation of collections of DNA fragments of interest; already in applied science, as a decisive element in itself, for example in clinical diagnosis.
Research
Conventional PCR is used as the basis for a multitude of techniques in the laboratory due to its robustness and speed. In this way, endpoint PCR makes it possible to control and detect the DNA fragments of interest.
An extremely important application of PCR is the cloning of DNA sequences into vectors, such as plasmids. For this, primers that contain at their 5' end are used. a short sequence that allows subsequent interaction with another complementary one located in the cloning vector to be used. For example, a restriction site can be included in said primers, so that, if this did not previously exist in the fragment and is unique in the vector, ligation can be carried out by T4 ligase after digestion with the T4 enzyme. appropriate restriction of both elements. Another method that can be assimilated to this route is the use of directed recombination; that is, it adapts to the 5' from the primers a sequence that enables a recombinase to target recombination with a given vector.
Medicine
In medicine, PCR is mainly used as a diagnostic tool (Coleman and Tsongalis, 2006):
- One of the main applications of the RCP is to do genetic testing to detect DNA mutations that cause some type of disease. The diagnosis of hereditary diseases present in the genome is a long and complicated process that can be significantly shortened by the RCP. Thus, DNA analysis of future parents can be done to see if they are carriers, or to analyze the DNA of their children if they are affected by hereditary disease. For prenatal analysis, DNA samples can be obtained from amniocentesis, from the sample of chorionic hairs or even from some fetal cells that circulate through the bloodstream of the mother. The use of RCP is also essential for the pre-plantation genetic diagnosis where embryo cells can be analyzed to look for possible mutations.
- It allows the genotyping of the species or species that cause a certain infectious picture: for this purpose, an area of the bacterial genome is amplified whose PCR product possesses some characteristics of size or fusion temperature that allow to identify it unequivocally. In the case of viral infections involving the integration of the pathogen genome into the host's DNA, such as HIV infection, the quantitative RCP allows the determination of the existing viral load and therefore the stage of the disease.
- RCP can also be used in routine medical checkups, such as in blood donor services; through this technique, donor infections (such as HIV or Hepatitis B) can be detected while still in the incubation period. Given the sensitivity of PCR tests, collective samples can be taken (e.g. 96 individual tests). If one of these collective samples is positive, progressively smaller samples are taken from it until the causator is found.
- The RCP can also be used as part of the tests performed when a tissue transplant is done. In 2008, it was proposed to use this technique to replace traditional antibodies testing.
- Sometimes, using the RCP you can do customized therapies for patients with certain types of cancer that produce mutations in the oncogenes from the detection of these mutations.
Paleontology, biological anthropology and forensic sciences
The fields of paleontology, biological anthropology, and forensic medicine and anthropology have been greatly benefited by this technique, since all of them frequently build knowledge of their corresponding disciplines thanks to remains or traces of living beings. One of the biological materials that can provide the most information is DNA.
The relative stability of this allows that, although fragmented, it is preserved for long periods if the conditions are propitious. Sometimes the intact samples that can be counted on are extraordinarily small or deteriorated. PCR solves both problems and provides useful amounts for subsequent analysis steps. In the first place, the amount of material recovered from scarce samples increases, since as previously stated, in theory a single molecule is enough for the process to take place. Also due to the nature of the technique and its purpose of amplifying small fragments, this fragmentation does not prevent this DNA from being used as a template for a PCR reaction.
- In paleontology and Anthropology the RCP allows to recover the few amounts of DNA that have not yet been degraded. Some places where DNA could be preserved are the brea volcanic ashes, the amber, historical polar ice or glaciers and arid environments, sediments, as well as in the apatite crystals of skeleton remains, being possible in that way to characterize corpses, fossils or other remains by genotyped by microsatellite analysis or even genome of excavaons, The purpose would be to use this amplified DNA to later carry out phylogenetic or ethnographic studies or populations by comparing DNA sequences, or by studying the causes of the evolutionary separation of two species.
- The forensic sciences are used to establish a person ' s filiation or to obtain evidence from minimal samples left by the perpetrator of a crime such as saliva, semen or other tissue remains (Butler, 2005).
Agronomy and diversity
Just as multiplex PCR makes it possible to produce genetic fingerprints of specific individuals, within the framework of forensic genetics, there are methods based on PCR that make it possible to discern between infraspecific groups of crops of agronomic interest; for example, from cultivars. To do this, oligonucleotides are used that are small enough to prime relatively nonspecifically, but always in such a way as to produce a discrete and interpretable banding pattern. In this way, the pattern obtained after the electrophoresis of the fragments tends to group the most similar individuals, who have a similar behavior, from which they diverge.
History
In 1971, an article published by Kleppe et al. in the Journal of Molecular Biology first described a method that used enzymes to replicate a small sequence of DNA with primers in vitro. However, this early example of the basic principle PCR did not receive much attention, and the invention of the polymerase chain reaction in 1983 is generally attributed to Kary Mullis. Mullis won the Nobel Prize for her work on PCR.
Something to take into account in PCR is that the DNA polymerase used is capable of withstanding the high temperatures of >90 °C necessary for the separation of the two DNA strands of the double helix after each cycle of replication. The DNA polymerases that were originally used for the in vitro experiments prior to PCR were not able to withstand these high temperatures, so the first procedures to replicate DNA were very inefficient, lengthy, and required large amounts of time. amounts of DNA polymerase.
The discovery in 1968 of Taq polymerase, a DNA polymerase extracted from the thermophilic bacterium Thermus aquaticus that inhabits very high temperature environments (50-80 °C), eliminated the major drawbacks of PCR method. This DNA polymerase is stable at high temperatures, remaining active until after DNA denaturation, eliminating the need to add new polymerase to the reaction after each cycle. This discovery made it possible to automate the process, previously so tedious, coupling it to the use of the thermal cycler.
At the same time that he was developing PCR in 1983, Mullis was working in Emeryville, California, USA, for an early biotech company, Cetus Corporation, where he was responsible for synthesizing short-stranded DNA. Mullis claims that he came up with the idea for PCR one night while driving his car across the Pacific Coast Highway. He was envisioning a new way to test for DNA mutations when he realized that instead of That, he had invented a method to amplify specific regions of DNA by repeated duplication cycles using DNA polymerases. Mullis attributes the invention of this technique to the effects of the hallucinogenic psychedelic drug LSD.
In the Scientific American magazine, Mullis summarized the procedure: "Starting with a single molecule of the genetic material DNA, PCR can generate 100 trillion identical molecules in an afternoon. The reaction is easy to do, requiring nothing more than a test tube, a few simple reagents, and a heat source". He was awarded the Nobel Prize in Chemistry in 1993 for his invention, and seven years later, he and his colleagues at Cetus put his proposal into practice.
Patent Wars
The PCR technique was patented by the Cetus Corporation, where Mullis worked when he invented the technique in 1983. The enzyme Taq polymerase was also covered by patents. Several lawsuits related to the technique took place, including an unsuccessful lawsuit filed by DuPont. The pharmaceutical company Hoffmann-La Roche acquired the rights to the patents in 1992 and currently maintains those that are still protected.
Contenido relacionado
Achromatic spindle
Cell (disambiguation)
Alkaloid
Commelinidae
Nightshade