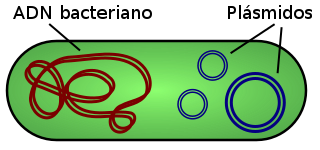
Plasmid
Plasmids are generally circular extrachromosomal DNA molecules that replicate autonomously and are transmitted (the latter by a process called conjugation) independently of chromosomal DNA. They are present mainly in prokaryotes (bacteria and archaea). In eukaryotes they are only found in yeast, but inactive plasmids have also been detected in mitochondria and chloroplasts, since these organelles are of bacterial origin. They are not infectious (pathogenic). They are examples of mobile genetic elements; Together with transposons and viruses, they constitute vehicles for horizontal gene transfer.
Plasmids are physically separated from the host chromosome. They are 3 to 10 kb in size and can carry 30 to 100 genes. The number of plasmids per cell can vary, depending on the type, from a single copy to several hundred. Plasmid vectors allow exogenous DNA ligands (outside the cell) of up to 4 kb to be cloned, since a size larger than this makes cloning in these vectors difficult.
Plasmids are acellular entities, thus they are similar to viruses and viroids in having the same replicative behaviour, transmission between hosts and being gene vectors. Although plasmids, viruses and viroids are "replicators" and evolve are generally not classified as living beings. Compared to viruses and viroids that depend on the host genome to replicate, plasmids replicate independently of the genome, therefore plasmids would fulfill one attribute of life: reproduction. Plasmids are phylogenetically related to DNA viruses such as Monodnaviria and some DNA satellite viruses since they both share a unique protein called Rep that allows for rolling circle replication and has no homology to cellular proteins., suggesting that plasmids and viruses may have origins that predate cells. It is also possible based on analysis of the Rep protein that some plasmids are precursors of the DNA viruses of the taxon Monodnaviria with the obtaining of proteins from certain viruses to make the capsid. Some plasmids could also have evolved from viruses of DNA according to phylogenetic analysis.
In contrast to viruses and viroids which are intracellular parasites, plasmids are mutualistic endosymbionts. Sometimes they establish beneficial relationships with their host cell while maintaining their independence in replication. However, on other occasions they can cause tumors. They carry antibiotic resistance genes. Although they are considered a genetic element of their hosts, they are not always inside them.
The term plasmid was introduced by the American molecular biologist Joshua Lederberg in 1952. The definition had to be refined over time, since it had initially been described in such a way that it included bacteriophages. Plasmids can only coexist as one or more copies in each bacterium, due to cell division they may be lost in one of the secreted bacteria.
They can be introduced into bacterial cells through a transformation process, which is why they are used as cloning vectors.
All cloning vectors must contain at least:
- A replication source to be able to have more than one copy of it in the infected cell.
- Two genes that confer resistance to different antibiotics (cloranfenicol and ampicillin), which allows the identification of cells that carry this vector.
Plasmid DNA molecules adopt a double-helical conformation just like the DNA of chromosomes, although, by definition, they are outside them. Plasmids have been found in almost all bacteria. Unlike chromosomal DNA, plasmids do not have associated proteins.
In general, they do not contain essential information, but confer advantages to the host under given growth conditions. The most common example is that of plasmids that contain genes for resistance to a certain antibiotic, so that the plasmid will only have an advantage in the presence of that antibiotic.
There are some integrative plasmids, that is, plasmids that have the ability to insert into the bacterial chromosome. These momentarily break the chromosome and are located inside it, with which the cellular machinery also reproduces the plasmid automatically. When that plasmid has been inserted they are given the name episome.[citation needed]
Plasmids are used as cloning vectors in genetic engineering because of their ability to reproduce independently of chromosomal DNA as well as because they are relatively easy to manipulate and insert new genetic sequences into.
Plasmids used in genetic engineering usually contain one or two genes that confer resistance to antibiotics and allow the selection of recombinant clones. There are other selection methods besides antibiotic resistance, such as those based on fluorescence or on proteins that kill cells without the use of antibiotics. These new plasmid selection methods are frequently used in agrobiotechnology, due to strong criticism from environmental groups for the possibility of antibiotics in genetically modified organisms.
Some plasmids include an addition system or postsegregational killing system (PSK, postsegregational killing system). These together produce a long-lived poison and a short-lived antidote. Daughter cells that retain one copy of the plasmid survive, while a daughter cell that fails to integrate the plasmid dies or suffers a reduced growth rate due to the plasmid. poison it got from the progenitor cell. This is an example of plasmids as replicating DNA.
From an evolutionary point of view, plasmids are considered selfish (or parasitic) genetic elements, since in many cases they do not share evolutionary interests with the chromosome of the cell in which they are found. Plasmids contain genes for their own maintenance and transmission, which can place an extra physiological burden on the cell. This phenomenon is known as bacterial fitness cost. However, plasmids can also confer highly beneficial functions to cells under certain conditions, such as antibiotic resistance genes in a selection environment. with these molecules. In addition, recent studies have shown that genes located on plasmids evolve in a different way than genes located on chromosomes. This suggests that the evolutionary implications of plasmids go far beyond being mere vehicles for transmission.
Episomes
An episome is a plasmid that can integrate itself into the chromosomal DNA of the host organism. For this reason, it can stay in contact for a long time, be duplicated at each host cell division, and become a basic part of its map. genetic. This term is no longer used in plasmids, since it is now clear that a region homologous to the chromosome makes a plasmid within an episome.
Plasmids used in genetic engineering are called "vectors." They are used to transfer genes from one organism to another and typically contain a genetic marker conferring a phenotype which can be selected for or against. Most also contain a polylinker or multiple cloning site (MCS), which is a small region that contains the most commonly used restriction sites, allowing easy insertion of DNA fragments at that site.
Types
One way to group plasmids is by their ability to transfer to another bacterium. Conjugative plasmids contain "tra-genes", which perform complex conjugation processes, such as sexual transfer of plasmids to another bacterium. Non-conjugative plasmids are incapable of initiating conjugation, hence they can transfer only with the assistance of conjugative plasmids and do so "by accident". An intermediate class of plasmids are "mobilizables" which carry only a subtype of genes required for transfer. They can "parasitize" a conjugative plasmid, transferring at a high frequency only in its presence.
It is possible for plasmids of different types to coexist in a single cell.
Seven different types of plasmids have been found in E.Coli. But normally related plasmids are incompatible, in the sense that only one of them survives in the cell line, due to the regulation of the vital functions of the plasmids. Therefore, plasmids can be differentiated according to compatibility groups.
Another way to classify plasmids is by function. There are 6 main classes:
Fertility Plasmids
Also known as F factors which contain tra-genes, they are capable of conjugation. It plays an important role with the conjugation of E. coli. In addition to being the first to be described, it is approximately 10 Kb long. It contains genes responsible for binding to the cell, and for the transfer of the plasmid located between specific bacterial strains in the conjugation process. Much of the information set for plasmid transfer is located in the tra operon, which contains less than 28 genes. These genes direct the formation of sexual pili that unite a donor cell with a recipient cell, other genes instead collaborate in the transfer of DNA. They also contain segments called insertion sequences, they assist in the insertion of the plasmid into the chromosome and into the host cell, so it can exist outside of the bacterial chromosome or be integrated into it.
Resistance Plasmids
Known as R factors, they confer resistance to certain antibiotics in hosts, they uniquely contain genes that code for enzymes capable of destroying or modifying antibiotics, they are not normally integrated into the chromosome of the bacterium that contains them, they have been found in the plasmids genes that encode resistance to antibiotics such as ampicillin, chloramphenicol and kanamycin, among others, some R plasmids contain a single resistance gene, others instead have up to 8, resistance genes are often found into a transposable element so that bacterial strains can rapidly develop plasmids encoding multiple resistances. Since many resistance plasmids are conjugation plasmids, they can spread throughout a population, although less rapidly than the fertility plasmid. Non-conjugative R factors often pass from one bacterium to another during conjugation promoted by another plasmid, with this method a whole population can become resistant to antibiotics. In fact, the fact that some of these plasmids can be easily transferred between species further promotes the spread of resistance. When the host consumes large amounts of antibiotics, bacteria with R factors are selected and become more prevalent, the R factors can then be transferred to more pathogenic genera such as Salmonella among others, causing greater public health problems. A good example of the importance of plasmids with antibiotic resistance genes in clinical settings is the high prevalence of the pOXA-48 plasmid in enterobacteria from hospitalized patients.
Col-plasmids
Bacteria also harbor plasmids with genes that give them a competitive advantage. In the world of microbes, bacteriocins are proteins that kill other bacteria, they can act only against closely related strains, or sometimes they kill cells by generating pores in them. the plasma membrane or degenerating the cell wall, thus causing its permeability to increase, another process to destroy the cell is degrading DNA and RNA or attacking peptidoglycan, col plasmids contain genes for the synthesis of bacteriocins known as colicins that are directed against e coli , there are plasmids with similar characteristics, which contain genes that encode bacteriocins directed against other species, for example, they produce cloacins that destroy enterobacter species, the The host is not affected by the bacteriocins it produces. Some Col plasmids are conjugative and contain resistance genes.
Degradative plasmids
These plasmids enable the digestion of unusual substances such as toluene or salicylic acid.[citation needed]
Virulent Plasmids
These plasmids turn the bacterium into a pathogen. They are capable of producing two types of toxins, a heat labile toxin (LT) which is a large protein very similar in structure and mechanism of action to cholera toxin, and a heat stable toxin (ST),
Metabolic Plasmids
They have genes for some rhizhobium strains to induce nodulation in legumes and carry out nitrogen fixation.
Conformations
Plasmid DNA can appear in one of five conformations, which (for a given size) run at different speeds on a gel during electrophoresis. The conformations are shown below in order of electrophoretic mobility (speed for a given voltage)., from slowest to fastest:
Circular open indentation
DNA has a single strand cut.
Linear
DNA has free ends, either because the strands were cut or because the DNA was linear in vivo. You can model this as a bead that hasn't connected to itself.
Circulate relaxed
DNA that fully interacts with both uncut strands, but has been enzymatically "relaxed." You can model this by leaving a bead relaxed and then connecting it to itself.
Denatured Super Coil
DNA like supercoiled or supercoiled DNA, but has unbound regions that make it slightly less compact; this results from excessive alkalinity during plasmid preparation.
Superspiral DNA
It is fully intact DNA with the strands uncut, and swirled, resulting in a compact shape.
The migration rate of small linear fragments is directly proportional to the applied voltage (in the case of low voltages). In the case of high voltages, large fragments continually migrate at different rates. Therefore, the resolution of the gel decreases with increasing voltage.
At a given low voltage, the migration of a small linear piece of DNA is a function of its length. Long (20kb) linear fragments migrate at a certain rate regardless of length. This is because the molecules "creep" from the center of the molecule following the direction of the gel matrix.
Restrictive digestion is frequently used to analyze purified plasmid fragments. These enzymes specifically break DNA at certain short sequences.
Applications
DNA Cloning
DNA cloning is a fundamental technique for obtaining large amounts of a specific DNA fragment. This fragment must first be joined to a vector DNA before being cloned. Vector DNA is a vehicle used to transport foreign DNA to a host cell, the vector contains sequences that allow it to replicate within this host cell.
In one technique, the segment of DNA to be cloned is inserted into a plasmid and then attached to a bacterial cell, at which point the bacterium captures the plasmid from the medium. In another alternative technique, the DNA segment is attached to a fragment of the lambdabacterial virus genome, then this virus infects a culture of bacterial cells, thus obtaining a large amount of virus, each virus containing of the foreign DNA fragment.
In either of the two techniques mentioned, once the foreign DNA fragment is inside the bacterium, it will be duplicated together with the bacterial or viral DNA, and distributed to the daughter cells. Thus the number of recombinant DNA molecules increases in proportion to the number of cells that are formed. So if we started with a single cell, in a short time we would have millions of copies of DNA. When the necessary number of copies is reached, the recombinant DNA can be purified and can be used in other processes. In addition to providing a means of amplifying the amount of particular DNA sequence, cloning can also be used as a technique for isolating any specific DNA fragment in pure form from a heterogeneous population of DNA molecules.
Cloning of eukaryotic DNA into bacterial plasmids
The foreign DNA to be cloned is introduced into the plasmid to form a recombinant DNA molecule. The plasmids used for DNA cloning are modified versions of those that can be seen in bacterial cells. In the same way their natural counterparts from which they are derived, have a duplication origin and one or more genes that confer antibiotic resistance to the recipient cell, antibiotic resistance allows selection of cells that contain the recombinant plasmid.
Bacteria that can take up DNA from a medium form a basis for cloning plasmids in bacterial cells. In this procedure, recombinant plasmids are added to a culture that has been pretreated with calcium ions. When the plasmid has been captured, it replicates autonomously inside the recipient cell. Bacterial cells containing the plasmid can be selected because they grow in the presence of the antibiotic against which they are to be resistant. When the desired amount of amplification is reached, the DNA is extracted, which can be easily separated from the recombinant DNA plasmid. In addition, isolated recombinant plasmids can be treated with the same restriction enzyme that was used in their synthesis, which releases the cloned DNA segments from the rest of the DNA that served as Vector. The cloned DNA can then be separated from the plasmid.
One of the main benefits of DNA cloning is that in addition to producing large amounts of a specific part of DNA, it allows different DNAs to be separated in a mixture. Initially it was noticed that bacteria that possess plasmids can be separated with antibiotic treatments, when this treatment is carried out they can be sedimented at low density in Petri dishes in such a way that the progeny of one cell remain separated from the progeny of another cell. Since there are a large number of recombinant plasmids, the different cells on the culture dish possess various foreign DNA fragments.
In the culture dishes that contain the bacterial colonies, the presence of a particular DNA sequence is investigated, using combined techniques of plate duplication and in situ hybridization. This duplication allows the preparation of a a large number of culture dishes containing representative colonies of the same bacterial cell, and are positioned in the same way in each dish. One of the replicas is used to locate a DNA sequence. This procedure requires lysing the cells and fixing the DNA on the surface of a filter. When the DNA is fixed, the DNA is denatured for hybridization. in situ, during which the filter is incubated with a labeled DNA probe that has the complementary sequence sought, later with the non-hybridity probe it is washed and the location of the labeled hybrids is determined by autoradiography, only then is selected living identified representatives of the cloning.
Although it is easy to search for a single human gene using this procedure, it is not a practical gene because it requires hundreds of prepared petri dishes.
Genetics
Plasmids serve as an important tool in genetics and biochemical engineering laboratories, where they are commonly used to multiply (make many copies of) or to express individual genes. Many plasmids are commercially available for such uses.
The gene to be replicated is inserted into copies of a plasmid which contain genes that make cells resistant to a particular antibiotic. In the next step the plasmid is inserted into the bacterium through a process called transformation. The bacteria is then exposed to a particular antibiotic. Only the bacterium that takes copies of the plasmid survives the antibiotic because the plasmid makes it resistant. In particular, protective genes are expressed (used to make protein) and the expressed protein prevents the action of the antibiotic. In this way, antibiotics act as a filter that selects only the modified bacteria. Now, these bacteria can be grown in large numbers, harvested, and the plasmid of interest isolated.
Another important use of plasmids is to make large amounts of proteins. In this, the bacterium containing the plasmid that encloses the gene of interest is allowed to grow. Just as the bacterium produces the protein that confers its resistance to antibiotics, it too can be used to produce proteins in large quantities from the inserted gene. This is a cheap and easy way to mass-produce genes or proteins that it encodes, such as insulin, or even antibiotics.
Plasmid DNA extraction
Much larger volumes of bacteria are grown in suspension in the maxiprep. Essentially, this is a scale-up of the mini-prep preparation, which is followed by further purification. This results in a relatively large amount (0.5 -1 mg) of very pure plasmid DNA.
In recent times many commercial kits have been created to perform plasmid extraction at various scales, purities and levels of automation.
Antibiotic resistance
Plasmids often contain genes or gene packages that give them a selective advantage, giving them the ability to make bacteria resistant to one or more antibiotics. Each plasmid contains at least one DNA sequence that serves as an origin of replication or ORI (an initial point for DNA replication), which enables DNA to be duplicated independently of chromosomal DNA.
Acquisition of genes needed to make these defense mechanisms is aided by a variety of mixed gene transfer systems, such as bacterial conjugative plasmids, transposable elements, and integron systems, which move genes around a system from DNA to another and from one bacterial cell to another, without the need for a relationship with the donor of the genes.
Bacterial plasmids serve as the scaffold on which antibiotic resistance gene arrays are assembled, by shuffling (transposable elements and ISCR-mediated shuffling) and site-specific recombination mechanisms (integron gene cassettes).
Contenido relacionado
Arundinella
Antinoria
Agrostis