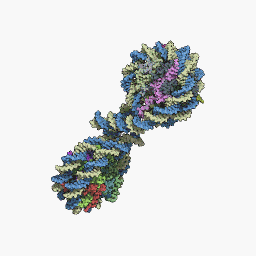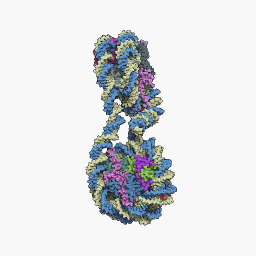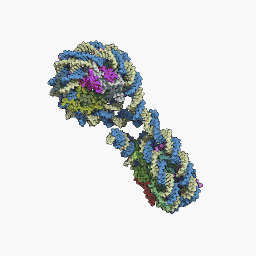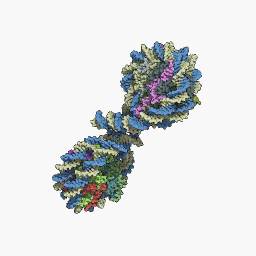
Nucleosome
The nucleosome is a structure that constitutes the fundamental unit of chromatin, which is the form of organization of DNA in eukaryotic cells, that is, the basic repeating unit of eukaryotic chromatin. A nucleosome is made up of approximately 145-150 base pairs of DNA supercoiled around a histone core, made up of two copies of each of the four histones. Nucleosomes are arranged like beads on a necklace, which, in turn, fold in on themselves repeatedly to form a chromosome. Histones are proteins rich in basic amino acids and highly conserved across the phylogenetic scale.
Discovery
In 1974, researchers Don and Ada Ollins were the first to observe nucleosomes, using electron microscopy, as 70 Å-diameter spherical particles in chromatin. In the same year, Roger Kornberg identified them and proposed their structure, based on the following in the following results:
- Chromatin contains approximately the same number of H2A, H2B, H3 and H4 histone molecules and no more than half H1.
- The H1 resides outside the nuclear particle of the nucleosome, it binds to the spacer DNA that connects a nucleosome particle to the next particle.
- X-ray crystallography indicated that in chromatin there is a regular structure that is repeated every 100 Å on the fiber. This same pattern is observed when purified DNA is mixed with equimolar amounts of histones, except H1.
- Electronic chromatin micrographs revealed that it consists of particles of approximately 100 Å of diameter connected by naked DNA (such as a bead necklace) and are apparently responsible for the X-ray pattern.
- Controlling the digestion time of chromatin with the microcoal nuclease (which breaks the double strand of DNA), fragments were obtained whose DNA always had multiple sizes of approximately 200 base pairs (quantified by electrophoresis experiments in gel).
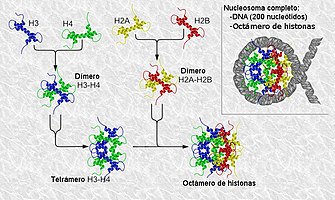
- Intercruzation experiments indicated that H3 and H4 histones are associated to form heterotetramer (H3)2(H4)2.
Kornberg's observations led him to conclude that the nucleosome consists of the octamer (H2A)2(H2B)2(H3)2(H4)2, plus about 200 base pairs of DNA. In direct contact with the center of the octamer are 140-147 base pairs. Between two adjacent octamers, the spacer DNA is 20–100 bp in length. Kornberg postulated that the fifth histone, H1, was somehow associated on the outside of the nucleosome.
When DNA is replicated in vivo, the daughter strands are immediately incorporated into nucleosomes. When DNA is replicated in the presence of cycloheximide (protein synthesis inhibitor), only one daughter strand has nucleosomes.
Later in 1987, researchers Lorch et al. demonstrated in vitro the role of nucleosomes as regulators of transcription and in 1988, Hans and Grunstein and Clark-Adams et al. demonstrated it in vivo.
Structure
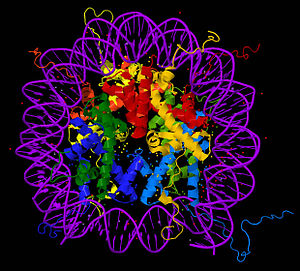
Nucleosome core particle
The core particle of the nucleosome is composed of a region of DNA 140-147 nucleotides in length, which wraps around the histone octamer in 1.7 turns in a negative or levorotatory supercoil. The octamer is made up of the core histones H2A, H2B, H3, and H4, each in pairs of two. Between each consecutive nucleosome there is a piece of DNA, called spacer DNA, whose length varies between 20 and 100 nucleotides depending on the length of the nucleosome. species and cell type. The entire structure forms a helix with a diameter of 11 nm and a height of 5.5 nm.
These particles can be observed by electron microscopy when the chromatin is in interphase and is forced to unpack by a partial DNase digestion. This generates a "bead or pearl necklace" shaped structure, where the string of the necklace corresponds to DNA and the beads/pearls to nucleosomes. DNase digestion of the longer regions of DNA accessible, those that are not bound to the nucleosome, generates fragments of multiple lengths of approximately 200 nucleotides. Using the gel electrophoresis technique, this fragmentation pattern can be observed. This type of DNA digestion can also occur under natural conditions during apoptosis, or programmed cell death.
N- and C-terminus
The extension of the N- and C-terminal tails of the histones accounts for up to 30% of their mass, but they are not visible in the structures of the nucleosomes obtained by crystallography due to their high intrinsic flexibility, so they are it is believed that they do not follow defined structures. The N-terminal ends of histones H3 and H2B are placed through the channels formed by the two strands of DNA, protruding from the DNA every 20 base pairs. On the other hand, the tail N-terminal of histone H4 contains a region of high content of basic amino acids (16-25), which form in the crystallographic structure interactions with highly acidic regions of the H2A-H2B dimer of other nucleosomes. This is potentially relevant to the structure of nucleosomes. These interactions are thought to occur under normal physiological conditions and suggest that acetylation of the H4 tail affects the upper structure of chromatin.
Structure of Chromatin
The repeat of the nucleosomes, in collaboration with the histone H1 in each other and with the spacer DNA, forms a compact 30 nm fiber, which reduces the length of the DNA by up to 50 times. For transcription to occur DNA, the DNA must be separated from the histones. In the unfolded state it is known as: Euchromatin. Euchromatin is currently believed to correspond to the 11 nm configuration and heterochromatin to the 30 nm configuration.
Contenido relacionado
Iridaceae
Magnoliidae
Muscat (grape)