Drosophila melanogaster
Drosophila melanogaster (Greek for literally "black-bellied dew lover"), also called vinegar fly or fruit fly, is a species of brachycerous diptera of the Drosophilidae family. It gets its name because it feeds on fruits in the fermentation process such as apples, bananas, grapes, etc. It is a species frequently used in genetic experimentation, since it has a reduced number of chromosomes (4 pairs), a short life cycle (15-21 days) and approximately 61% of the known human disease genes have an identifiable counterpart. in the genome of fruit flies, and 50% of the fly protein sequences have mammalian analogues.
For research purposes, they can easily replace humans. They reproduce quickly, so that many generations can be studied in a short space of time, and the complete map of their genome is already known. It was adopted as a genetic experimental animal by Thomas Morgan in the early 20th century. Its 165 Mb of genome (1 Mb = 1 million base pairs) were published in March 2000 thanks to the public consortium and the company Celera Genomics. It houses around 13,600 genes.
Development
From a cell derive daughter cells that generate a possible asymmetry. It presents an initial asymmetry in the distribution of its cytoplasmic components that gives rise to its developmental differences. In oogenesis, follicular cells, nurse cells and the oocyte are generated. The fruit fly, at 29 °C, manages to live 30 days; and development from egg to adult requires 7 days.
The primordium develops differences in the axes: anteroposterior and dorsoventral.
A succession of events derived from the initial asymmetry of the zygote results in the control of gene expression in such a way that different regions of the egg acquire different properties. This may occur due to the different localization of the transcription and translation factors in the egg or due to the differential control of the activities of these factors.
This is followed by another stage in which the identities of the parts of the embryo are determined: regions are defined from which specific parts of the body are derived.
Genes that regulate the process encode transcription regulators and act on each other in a hierarchical fashion. They also act on other genes that are truly responsible for establishing this pattern (they act in a cascade).
Cell-cell interactions must also be taken into account as they define the boundaries between cell groups.
Structure of a segment
There are 3 groups of genes based on their effects on the structure of a segment:
- Genes maternos: expressed by the mother in the ovogenesis. Act during or after the ripening of the egg. An example is the bicoid gene.
- Segmentation genes: expressed after fertilization. They are responsible for the number and polarity of the segments (there are 3 groups that act sequentally to define the parts of the embryo).
- Genes homeotics: control the identity of the segments (not number, polarity or size).
Stages of development
The next stage of development depends on the genes that are expressed in the parent fly. These genes are expressed before fertilization. They can be divided into:
- Maternal Somatic Genes: are expressed in somatic cells = follicular cells.
- Maternal germline genes: they can act in both rot cells and oocyte.
There are four groups of genes involved in the development of different parts of the embryo. Each group is organized in a different way that presents a specific order of action. Each pathway begins with events that take place outside the egg, resulting in the location of a signal within the egg. These signals (these are proteins called morphogens) are distributed asymmetrically to fulfill different functions.
Three systems are in charge of the anteroposterior axis and one is in charge of the dorso-ventral:
- Previous system: responsible for the development of head and chest. Maternal germline products are required to place the bicoid gene product at the previous end of the egg.
- Post-: responsible for the segments of the abdomen. Many products intervene in the location of the nano gene product, which inhibits the hunchback expression in the abdomen.
- Terminal system: development of structures of the non segmented ends of the egg. It depends on mathematical somatic genes (activate the torso-coded receptor).
- Dorso-ventral system: starts by a signal from a follicular cell of the egg ventral face and is transmitted through the receptor encoded by the Toll gene. This produces the generation of a gradient of activation of the transcription factor produced by the Dorsal gene.
All the components of the four systems are maternal, so the systems that establish the initial pattern depend on events prior to fertilization.
Dorso-ventral development
There is a complex interrelationship between oocyte and follicular cells (oocyte genes are necessary for the development of follicular cells and signals from these, transmitted to the oocyte, cause the development of ventral structures).
Another pathway is responsible for dorsal development during egg growth.
The systems work by activating a ligand-receptor interaction that triggers a transduction pathway.
The process depends, at its beginning, on the Gurken gene (which also acts in anterior-posterior differentiation). Gurken mRNA localizes to the posterior face of the oocyte, causing adjacent follicular cells to differentiate into posterior cells. These cells return a signal that triggers the production of a microtubule network that is necessary for polarity.
Dorsoventral polarity is established when gurken reaches the dorsal face of the oocyte (depends on the expression of several other genes).
The Gurken product acts as a ligand interacting with the receptor (product of the Torpedo gene) of a follicular cell.
Activation of this receptor triggers a signaling pathway whose final effect is to prevent the development of the ventral face into the dorsal face (a change is produced in the properties of the follicular cells on this face).
The development of ventral structures requires maternal genes that establish the dorso-ventral axis. The dorsal system is necessary for the development of ventral structures (such as mesoderm and neuroectoderm). Mutations in it prevent ventral development.
The ventral developmental pathway also begins in the follicular cells and ends in the oocyte. In the follicular cells, a series of signals are produced that end up generating a ligand for the receptor (product of the Toll gene = first component of the pathway, which acts inside the oocyte).
Toll is the crucial gene in the transport of the signal inside the oocyte.
The rest of the components of the dorsal group encode products that either regulate or are necessary for the action of Toll. Toll is a transmembrane protein (homologous to the interleukin 1 receptor).
The binding of its ligand to the Toll receptor activates the pathway that determines ventral development. The distribution of the product of this gene is highly variable, but it only induces the formation of ventral structures in suitable places (it seems that the active product is only expressed in certain regions).
After ligand binding, the Toll receptor is activated on the ventral side of the embryo. This activation triggers a series of processes in which the products of other genes are involved and which ends in the phosphorylation of the product of the cactus gene, which is the final regulator of the transcription factor of the Dorsal gene.
In the cytoplasm there is an inactive cactus-dorsal complex, but when cactus phosphorylates, it releases the dorsal protein, which enters the nucleus.
Activation of toll leads to activation of dorsal.
A dorsal protein gradient is established in the nucleus running from the dorsal to the ventral side in the embryo. On the ventral side, the dorsal protein is released into the nucleus but on the dorsal side, it remains in the cytoplasm.
The dorsal protein activates the Twist and Snail genes (necessary for the development of ventral structures) and inhibits the Decapentaplegic and Zerknullt genes (necessary for the development of dorsal structures). The initial interaction between gurken and torpedo leads to repression of spatzle activity on the dorsal side of the embryo (toll ligand).
The dorsal protein, located in the nucleus, inhibits the expression of dpp. Thus, the ventral structures are formed along a nuclear gradient of the dorsal protein and the dorsal structures along a gradient of the dpp protein.
- In the dorso-ventral axis there are three rather nearby bands that define the regions in which mesoderm, neuroectoderm and dorsal ectoderm are formed (ventral to dorsal orders).
History of use in genetic analysis
D. melanogaster was one of the first organisms used for genetic analysis, and is today one of the most widely used and genetically best known eukaryotic organisms.
Thomas Hunt Morgan began using this species at Columbia University in 1910 in a laboratory known as "the fly room". He and his collaborators (including the famous geneticists A.H. Sturtevant, Calvin Bridges, and H. J. Muller), began experiments using milk bottles to raise flies and magnifying glasses to observe them. The magnifying glasses were later replaced by dissecting microscopes. Using these small, harmless flies, Morgan and his colleagues elucidated many basic principles of heredity, including sex-linked inheritance, epistasis, multiple alleles, and gene mapping.
Within the genetic analysis, loss-of-function studies were carried out, which implied the silencing of various genes to observe what function they carried out. In this way, the fruit fly genome was altered, firstly, through the use of mutagens that caused changes in its DNA sequence without strict experimental control, such as transposable genetic elements, X-rays or mutagenic chemicals. Later on, RNA interference (RNAi) was used which, together with the GAL4-UAS system, allows editing in a specifically determined tissue. Lastly, and most recently, its genome is studied using the CRISPR system together with Cas9 nuclease, allowing easier, more effective and affordable editing. Thanks to this discovery, we have proceeded to develop new nucleases with complementary functions to Cas9 that allow further editing and study of this organism through this system, such as the Cas12a nuclease (previously known as Cpf1), and the CasΦ nuclease.
Behavioral genetics and neuroscience
Seymour Benzer and others have used mutations that affect the behavior of these flies to isolate genes involved in vision, smell, hearing, learning, memory, courtship, pain, and other processes.
Following the pioneering work of Alfred Henry Sturtevant, Benzer and colleagues used gynandromorphs (sex mosaics) to develop the new fate mapping technique. This technique made it possible to assign a particular feature to a specific anatomical location. For example, this technique demonstrated that male sexual courtship behavior is controlled by the brain.
Mapping the fate of gynandromorphs also provided the first indication of the existence of pheromones in this species. Males distinguish between conspecific males and females and direct courtship towards females, thanks to a specific sex pheromone that females produce mainly on their legs. tergites (hardened dorsal plates on the outside of the abdomen).
Genome
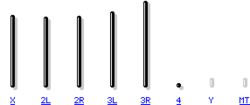
The genome of D. melanogaster (sequenced in 2000, and verified in the FlyBase database) contains four pairs of chromosomes: one X/Y pair, and three autosomes labeled 2, 3, 4. The fourth chromosome is so small that it is sometimes ignored, except for the important eyeless gene. The sequenced genome of D. melanogaster of 139.5 million base pairs contains approximately 15,016 genes. More than 60% of its genome is functional, encoding non-coding DNA for proteins involved in the control of gene expression. Sex determination in Drosophila is produced by the relationship of X chromosomes to autosomes, not due to the presence of a Y chromosome as occurs in sex determination in humans. Although the Y chromosome is entirely heterochromatic, it contains at least 16 genes, many of which have male sex functions.
Similarity to humans
About 75% of human disease-related genes have their homologues in the fruit fly genome, and 50% of the fly protein sequences have their mammalian homologues. An online database, called Homophila, is available for studies of human genetic disease homologs in flies and vice versa. Drosophila continues to be widely used as a genetic model for various human diseases including neurodegenerative disorders Parkinson's, Huntington's, spinocerebellar ataxia and Alzheimer's. This fly is also used in studies of mechanisms of aging and oxidative stress, the immune system, diabetes, cancer, drug abuse.
Contenido relacionado
Achromatic spindle
Cell (disambiguation)
Alkaloid
Commelinidae
Nightshade