Domain (biology)
In biology, the domain is a taxonomic category used in biological classification systems, above kingdom and superkingdom. The domain has been added above the kingdoms to place in one category the three taxa: archaea (Archaea), bacteria in the narrow sense (Bacteria sensu stricto) and eukaryotes (Eukarya); while the category of empire or superkingdom was added by taxonomists who needed a category above the kingdoms to place prokaryotic (or Bacteria in the broad sense) and eukaryotic taxa in it.
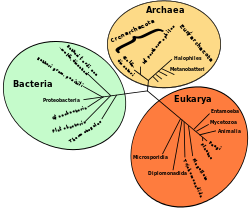
The three domains were proposed by Carl Woese in 1990 when he created, applying the new cladistic taxonomy, his three-domain system. He proposed the three domains on the basis of a phylogenetic tree hypothesis in which these 3 clades diverged from an ancestral parent. Alternatively, in a traditional evolutionary system, living beings are classified into a System of two super-kingdoms, or, if the category of empire is used, of two empires: Prokaryota and Eukaryota; The name of the two groups alludes to the presence or absence of a cell nucleus, a character accompanied by a significant number of characters that define each group. Today the prokaryotic group can be considered to be paraphyletic, since the eukaryotic domain originates from prokaryotic ancestors.
Viruses are totally dependent biological systems, parasites, that many scientists prefer to consider as non-living, so they are classified separately. For them, the taxonomic category Acytota (acellular) has been proposed. According to phylogenetic studies by the International Committee on Taxonomy of Viruses, viruses comprise more than 6 domains.
| Two superrenants (Chatton) 1925, 1938) | Three domains (Woese) 1977, 1990) |
|---|---|
| Eukaryota | Eukarya |
| Prokaryota | Archaea |
| Bacteria |
Three domain system
The new biological studies were developed at the molecular level, phylogenetically, 16s ribosomal RNA sequencing showed that within prokaryotes, archaea were fundamentally different from bacteria as well as eukaryotes.
In the Archaea and Bacteria domains only unicellular organisms are included, in their morphology they appear to be simple and not very diverse, but with a great variety of metabolisms and nutritional dependencies. All organisms with complex anatomy, along with other simpler or unicellular ones, belong to the Eukarya domain (the eukaryotes), which includes the animal, plant, fungal, and protist kingdoms.
Criticism
Many authors, especially evolutionary biologists, do not accept the three-domain system; although it is commonly incorporated into Archaea and Bacteria as prokaryotes. Woese's hypothesis is based on some genetic aspects, but ignores aspects of life history, genetic structure, ecology, symbiotic relationships, morphology, and evolutionary development (Margulis 1982).
As has been shown (Cavalier-Smith 2002a, 2006a,c), ignoring the structure of organisms, cell biology, and paleontology led to a now widespread fundamental, but misleading, interpretation of history of life, the system of three domains. This serious error is based not only on not integrating the sequence evidence with other data, but also on not taking into account the often extremely non-clock nature of sequence evolution, and on systematic errors that strongly guided other conclusions, in sequence trees for molecules that do not evolve according to naive statistical preconceptions (see Cavalier-Smith 2002a, 2006a, c). Transition analyzes using complex three-dimensional characters are less prone to phylogenetic artifacts than sequences, and provide powerful evidence that the tree topology is very different (see Cavalier-Smith 2002a, 2006a, c, Valas and Bourne 2009).). Paleontology provides equally compelling evidence that cyanobacteria are substantially older than eukaryotes (Cavalier-Smith 2006a). The statistics used in the phylogeny analyzes failed to adequately model the extremely rare single historical events of megaevolution, for which the uniformity assumptions are entirely invalid (Cavalier-Smith 2006b). (in Cavalier-Smith 2010)
System of two domains or superkingdoms
This system divides living beings into two large groups: prokaryotes and eukaryotes. This dichotomy is based on the great differences between them, considering that they represent the greatest evolutionary discontinuity in the history of the Earth. The differences are at all levels: in complexity, size, morphology, ecology, cell structure, reproduction, symbiotic relationships, evolutionary development, biochemistry and genetics (of DNA, RNA and proteins).
The apparent equidistance between the groups Eukaryota, Archaea and Bacteria, is actually due to the complex process that gave rise to eukaryotes, eukaryogenesis, which was produced by symbiogenesis between an archaea and a bacterium; that is, eukaryotes are descendants of prokaryotes and are also much later than these, consequently they have inherited characteristics of both Archaea and Bacteria. The eukaryotic genetic revolution is very abrupt and therefore gives the impression that it was much older, as explained by the attraction factor of long branches.
Probably new domain
In 2012, in hydrothermal vents in Japan, the discovery of Parakaryon myojinensis was carried out, a unicellular organism that presents characteristics that do not fit with the cells of the other three domains and probably constitutes its own domain Parakaryota. Parakaryon myojinensis, like eukaryotes, has a nucleus and other endosymbionts in its cell, however its nuclear envelope is a single layer, not made of two concentric membranes as in any eukaryote, and the genetic material is stored as in bacteria, in filaments and not in linear chromosomes. In addition, it does not present endoplasmic reticulum, Golgi apparatus, cytoskeleton, mitochondria, nuclear pores and totally lacks flagellum. Ribosomes are found not only in the cytoplasm but also in the nucleus. It has a cell wall composed of peptidoglycans like bacteria and its mode of nutrition is osmotrophic. It is difficult to specify its phylogenetic relationship with other living beings because only a specimen has been found and its genome could not be sequenced. According to some authors it would be an intermediary organism between prokaryotes and eukaryotes.
Supergroup system
Another taxonomic category that replaces kingdoms, domains and empires is that of supergroups as used for example in Adl et al. (2005).
Historical aspects
Several authors have theorized a taxonomic category higher than that of the biological kingdoms. Some outstanding systems are the following:
In 1735 Charles Linnaeus created the taxonomic classification system and used the term "empire" to refer to nature, the source of his study: the Imperium Naturae divided into the regnum Animalia , Vegetabilia and Lapides (ore).
The prokaryotic concept has its oldest equivalent in Ferdinand Cohn, who in 1875 gathered the smallest beings: bacteria and blue-green algae into a single group, called it Schizophyta and placed it within the Plant kingdom.
Chatton introduced the terms “prokaryote” and “eukaryote” in 1925, to differentiate nucleated from anucleated microorganisms. In 1938 he included plants and animals among eukaryotic beings. Actually, Chatton was not referring directly to a classification with taxonomic levels higher than the kingdoms; however he laid the foundations of the most well-known and fundamental biological dichotomy. The French biologist André Lwoff, a disciple of Chatton and later the Canadian microbiologist Roger Stanier, spread this categorization of living beings since the middle of the XX< century.
A notable advance in prokaryotic phylogenetic study was ribosomal analysis. In 1977 it was discovered that the greatest difference according to rRNA is between archaea and bacteria. The comparison between these and eukaryotes allowed making a proposal in three primary kingdoms: "eubacteria, archaebacteria and urkaryotes", which forms the basis of the current three-domain system.
Virus situation
Viruses were technically discovered in 1899 when the Dutch microbiologist Beijerinck defined them as an infectious agent that only multiplies inside living dividing cells, but since his experiments did not show that it was composed of particles, he called it contagium vivum fluidum («living soluble germ») or «virus». In 1930 the Czech botanist F.A. Novák defines them as a group of organisms not made up of cells with the name of Aphanobionta and in 1931 the first images were obtained with the invention of electron microscopy.
In 1957 André Lwoff defined the main differences between viruses and bacteria based on molecular structure and physiology: "Viruses must be treated as viruses, because viruses are viruses." There are no biological entities that can be described as transitional between a virus and a cellular organism, and the differences between them were of such a nature that it is really difficult to visualize any kind of intermediate organism. In 1962 Lwoff proposed the first taxonomic classification for viruses, which goes far apart from cellular organisms.
Most biologists exclude viruses from biological classification systems. According to the cell theory, viruses are not living beings, although they only have the capacity to evolve like cells. Philosophically, it has been argued that viruses, along with subviral agents, are mobile genetic elements due to their replicative behavior and because they are vectors. of genes; thus they are more comparable to plasmids than to cellular life forms. Another important factor is that the viruses did not meet the necessary requirements to be able to reliably locate them in the tree of life: for example, they do not have ribosomes, they all lack a nucleic acid, they have no fossil record, and there is not even a shared gene. Among the majority of viral groups, since they are newly synthesized, most of their genes are mixed with those of their hosts and, as a last resort, the viral particles do not have a common ancestor, so they are a polyphyletic set. has decided to compare viruses with mobile genetic elements or cellular sequences in phylogenetic analyses.
According to phylogenetic studies carried out by the International Committee on Taxonomy of Viruses, viral entities can be divided into 7 domains based on their protein or molecular characteristics, Riboviria which includes RNA viruses and reverse transcription, Ribozyviria including virusoids, Viroidia including viroids, Adnaviria, Duplodnaviria, Varidnaviria and Monodnaviria which include only DNA viruses, these domains in turn comprise more than 10 kingdoms. In addition, there are plans to create more domains that include unclassified DNA viruses and subviral agents (unclassified satellite viruses). Since viruses and subviral agents are not accepted to be classified with cellular organisms, ICTV uses the term &# 34;realm" for domain, which does not have a fixed translation into Spanish, since they wanted to avoid confusion with cell life domains.
When viruses are included together with cellular organisms, for example: in protein analyses, viruses branch in a complexly paraphyletic manner with respect to the three cellular domains, even using more realistic evolutionary models such as maximum parsimony. Large DNA nucleocytoplasmic viruses were closest to cellular organisms, followed by other DNA viruses and RNA viruses along with reverse transcription viruses, with the family Mimiviridae being the sister group. of cellular organisms. Because viral proteins have no homologues with cells and some unique genes have been conserved in certain viral lineages, it has been suggested that viruses originated during the RNA world in protobionts that diverged to give rise to prokaryotic cells. Several viruses infecting archaea and bacteria would have already infected the last universal common ancestor and protobionts. A recent hypothesis proposes that viruses, subviral agents, mobile genetic elements, enzymes, cellular sequences, etc. were the replicons of the RNA world and ancestors. of cellular genetic components. Viruses may have served to mediate horizontal gene transfer between self-replicating communities of protobionts.
The various historical domain-based systems (including viruses) can be summarized in the following table:
| Linnaeus 1735 1 empire | Chatton 1925 2 groups | Novák 1930 3 groups | Allsop 1969 2 superrenants | Jeffrey. 1971 3 superreinos | Woese 1990 3 domains | BioLib 2008 5 domains | Ruggiero 2015 2 superrenants |
|---|---|---|---|---|---|---|---|
| Naturae | Eukaryote | Karyonta | Eucaryotae | Eucytota | Eukarya | Eukaryota | Eukaryota |
| Procaryote | Akaryonta | Procaryote | Procytota | Bacteria | Bacteria | Prokaryota | |
| Archaea | Archaea | ||||||
| Aphanobionta | Acytota | Nucleacuea | |||||
| Aminoacuea |
Contenido relacionado
Kalanchoe
Dieffenbachia
Sageraea