Dna viruses
A DNA virus is a virus whose genetic material is made up of DNA, not using RNA as an intermediary during replication. Viruses that use RNA either as genetic material or as an intermediary during replication are RNA viruses. DNA can be both single-stranded (single-stranded) and double-stranded (double-stranded), the latter being more diverse and frequent. Replication within cells depends on a DNA-dependent DNA polymerase (which reads the DNA). It is common for single-stranded DNA to expand to double-stranded DNA in infected cells.
Baltimore Classification and Replication
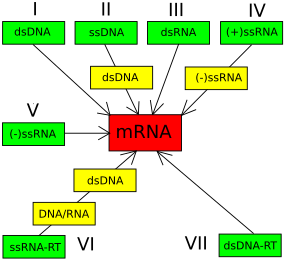
The Baltimore classification system is used to group viruses based on their mode of messenger RNA (mRNA) synthesis, and is often used in conjunction with standard virus taxonomy, which is based on evolutionary history. DNA viruses constitute three Baltimore groups: Group I: double-stranded DNA viruses, Group II: single-stranded DNA viruses, and Group VII: double-stranded reverse-transcribed DNA viruses. Although the Baltimore classification is based primarily on mRNA transcription, the viruses in each Baltimore group also often share their mode of replication. The viruses in a Baltimore cluster do not necessarily share a genetic or morphological relationship.
Double-stranded DNA Viruses
The first group of Baltimore DNA viruses are those with a double-stranded DNA genome. All double-stranded DNA viruses have their mRNA synthesized in a three-step process. First, a pretranscriptional initiation complex binds to DNA upstream of the site where transcription begins, allowing for the recruitment of a host RNA polymerase. Second, once RNA polymerase is recruited, it uses the negative strand as a template to synthesize the mRNA strands. Third, RNA polymerase terminates transcription upon reaching a specific signal, such as a polyadenylation site.
Double-stranded DNA viruses use several mechanisms to replicate their genome. Bidirectional replication, which is the typical form of DNA replication in eukaryotes, is widely used. In bidirectional replication, a circular genome is cleaved to separate the two strands, creating a bifurcation from which replication of both strands progresses around the genome at the same time, going in two opposite directions until reaching the opposite end. A rolling circle mechanism is also used which produces linear strands while looping around the circular genome equally replicating both strands simultaneously. Instead of replicating both strands at once, some double-stranded DNA viruses use a strand displacement method whereby one strand is synthesized from a template strand and then a complementary strand is synthesized from the previously synthesized strand, forming a double-stranded DNA genome. Finally, some double-stranded DNA viruses replicate as part of a process called replicative shuffling whereby a viral genome in one host cell's DNA is replicated in another part of a host genome.
Double-stranded DNA viruses can be subdivided into those that replicate in the nucleus and, as such, are relatively dependent on the host cell's machinery for transcription and replication, and those that replicate in the cytoplasm, in which case they have evolved or acquired their own means to execute transcription and replication.
Single-stranded DNA viruses
The second group of Baltimore DNA viruses are those with a single-stranded DNA genome. Single-stranded DNA viruses have the same form of transcription as double-stranded DNA viruses. However, because the genome is single-stranded, it is first converted to a double-stranded form by a DNA polymerase upon entering a host cell. The mRNA is then synthesized from the double-stranded form. The double-stranded form of single-stranded DNA viruses can occur directly after entry into a cell or as a consequence of replication of the viral genome. Eukaryotic single-stranded DNA viruses replicate in the nucleus.
Most single-stranded DNA viruses contain circular genomes that replicate by rolling circle replication (RCR). RCR single-stranded DNA is initiated by an endonuclease that binds to and cleaves the positive strand, allowing a DNA polymerase to use the negative strand as a template for replication. Replication progresses in a loop around the genome by extension of the 3' end of the plus strand, displacing the previous plus strand, and the endonuclease cleaves the plus strand again to create an independent genome that ligates in a loop circular. The new single-stranded DNA can be packaged into virions or replicated by DNA polymerase to form a double-stranded form for transcription or continuation of the replication cycle.
Parvoviruses contain linear single-stranded DNA genomes that replicate by rolling hairpin replication (RHR), which is similar to RCR. Parvovirus genomes have hairpin loops at each end of the genome that repeatedly unfold and refold during replication to change the direction of DNA synthesis and move back and forth throughout the genome, producing numerous copies of the genome. genome in a continuous process. Individual genomes are then excised from this molecule by viral endonuclease. For parvoviruses, either the positive or negative sense strand can be packaged into capsids, which vary from virus to virus.
Almost all single-stranded DNA viruses have positive-sense genomes, but there are some exceptions and peculiarities. The family Anelloviridae is the only single-stranded DNA family whose members have negative-sense genomes, which are circular. Parvoviruses, as mentioned above, can package either the positive or negative sense strand into virions.
Reverse transcribed double-stranded DNA virus
The seventh Baltimore group contains viruses that have a double-stranded DNA genome that replicate by an RNA intermediate in their replication cycle. Double-stranded reverse transcribed DNA viruses have a gap in one strand, which is repaired to create a complete reverse transcribed double-stranded DNA genome prior to transcription. Double-stranded reverse-transcribed DNA viruses transcribe in the same way as double-stranded DNA viruses, but make use of reverse transcription to replicate their circular genome while still in the capsid. The RNA polymerase II of the host cell transcribes the RNA strands of the genome in the cytoplasm, and the genome is replicated from these RNA strands. The reverse transcribed double-stranded DNA genome is produced from pregenomic RNA strands by the same general mechanism as reverse transcribed single-stranded RNA viruses, but replication occurs in a loop around the circular genome. After replication, the reverse transcribed double-stranded DNA genome can be packaged or sent to the nucleus for further rounds of transcription.
Origin and evolution
According to the International Committee on Taxonomy of Viruses (ICTV) DNA viruses can be found in five of the six established domains and could have diverse evolutionary origins.
- Duplodnaviria: Contains DNA viruses that have a specific protein called HK97-MCP and a single enzyme ends it within the caucus. They are bicatenary DNA viruses and icosaedric capsides. It includes the classic caudovirus of procariots and their eukaryotic descendants; herpesvirus and mirusvirus. It has been suggested that they arose before the last universal common ancestor (LUCA) and that the caudovirus preceded the herpesvirus and mirusvirus with the loss of the contractable tail. The fusion of a precellular DNA replication with a polymerase DNA, the ending, the HK97-MCP and DUF1884 domain proteins, helice-hebra, dodecins and encapsulins led to the formation of the virions Duplodnaviria, that at first they would not have the contractable tail, so it would have developed after LUCA.
- Adnaviria: Includes archaeal viruses containing a specific dimer protein called SIRV2 inside the capside, filamentous morphology and A-shaped DNA. All representatives are bicatenary DNA viruses. Its origin is prior to the last universal common ancestor (LUCA) due to the lack of cellular counterparts. The fusion of a precellular DNA replication with polymerase DNA, SIRV2 and propeller-herited domain proteins, METT26, SER2, ILE26 and ARG128 led to the formation of the virions Adnaviria. It is also the only domain that so far has no descendants in the eukaryotes.
- Varidnaviria: Contains DNA viruses that have proteins in vertical jelly roll can be in double DJR-MCP roll or in simple SJR-MCP roll within the capside. Almost all of them are from bicatenary DNA, except a family that is from monocatenary DNA, but they evolved from bicatenary DNA viruses from this domain. The members of this domain have mostly icosaedrica capsides, although in some members the icosaedrica capside has evolved into an ovoid or brick form. It is divided into two kingdoms: Bamfordvirae which includes viruses with proteins in double roll of jelly DJR-MCP among which are adenovirus, giant viruses and various procarotes such as tectivirus or turrivirus, while Helvetiavirae includes the viruses with the simple roll jelly proteins SJR-MCP that includes procarote viruses that appear to have given rise to the viruses Bamfordvirae by joining two SJR-MCP in a single fold originating the DJR-MCP. It has been suggested that the origin of Varidanviria It is also prior to the last universal common ancestor (LUCA) and that the eucariovirus of the domain descends from tectivirus through intermediate viruses called "polintovirus" which would later become the polynton transposons. The fusion of a precellular DNA replication with polymerase DNA and cupin domain proteins, TNF, Pro-PD, CBM, nucleoplasmins and vertical SJR-MCP led to the formation of the virions Varidnaviria. Giant viruses evolved from small viruses of polyntovirus, which increased their genome and the size of the virion by duplicating and deletion of genes, including mobile genetic elements and massive acquisition of host genes and their endosymbiotic bacteria, including genes for translation and computer genes that are considered to be the most resistant to horizontal transfer. Giant viruses may have given rise to unusual class viruses Naldaviricetes or descend from a shared ancestor.
- Monodnaviria: Contains DNA viruses with a specific protein called REP that makes a replication in a rolling circle and an HUH endonuclease. It includes almost all monocatenary DNA viruses along with papilomavirus and polyomavirus that are of bicatenary DNA, but that evolved from monocatenary DNA viruses. Members of this domain have icosaedric capsides, although some members have helical capsides. It is divided into four kingdoms Shotokuvirae which includes the eucariovirus of the domain such as papilomavirus, poliomavirus, circovirus, parvovirus, nanovirus, geminivirus, etc. The other kingdoms Sangervirae, Trapavirae and Loebvirae, they are composed exclusively of procarote viruses, Loebvirae stands out for including filamentous bacteriophages such as inovirus, while others include icosaedric bacteriophages without tail such as microvirus. It has been suggested that the viruses of this domain arose through three events in which plasmids were integrated into the positive monocatenary RNA virus capsides, replacing the genome and the typical enzymes of the virus. The eucariovirus of the domain (Shotokuvirae) descend from bacteriophages related to microvirus with which they are more closely related and share structural characteristics. Viruses Monodnaviria appear to have originated the superfamily of helitron transposons and some types of plasmids.
- Fusiform viruses and Clavaviridae: It has been suggested that families of fusiform viruses or in the form of archaic lemon Fuselloviridae, Halspiviridae, Thaspiviridae and Bicaudaviridae are related by the presence of a homologous protein SSV1, ATPasas de la superfamilia AAA, forma de lemon y el assemblage, the homologa protein is also present in the family of arqueanos filamentososos virus Clavaviridae, so it has been suggested that these viruses evolved from filamentous viruses related to Clavaviridae that altered their form to store a larger genome and would form a domain or lineage. Its origin as well as other domains is prior to the last universal common ancestor (LUCA).
- Riboviria: is a domain that includes all RNA viruses and retrotranscribed viruses so it includes retrotranscribed bicatenary DNA viruses even though all other viruses are RNA. These are characterized by having the reverse transcriptase enzymes and the RNA-dependent polymerase RNA (RdRP) that was believed to be the most primitive enzymes that transcribe nucleic acids. The retrotranscribed bicatenary DNA virus that contains two families the hepadnavirus and the caulimovirus that infect animals and plants are included in the kingdom Pararnavirae along with the retrotranscribed monocatenary RNA viruses. It has been suggested that these two groups originated from an event in which a non-LTR retrotranspoponse was integrated into the cpside of a RNA virus, replacing the genome and the typical enzymes of the virus and that the retrotranscribed bicatenary DNA viruses are polyphiletic being the caulimovirus a branch derived from the retrotranscribed monocatenary RNA virus and the basal hepad. The retrotranscribed viruses could emerge a bit after the eukaryotes originated.
In addition, there are about 16 families of DNA viruses not assigned to any domain, which are archaeal double-stranded DNA viruses with unusual capsids that are difficult to classify due to the lack of homology with other DNA viruses, therefore they are considered to be disconnected from the virosphere. DNA viruses with unusual capsids from archaea have been suggested to be descendants of several extinct lineages of DNA viruses that existed before the last universal common ancestor (LUCA). Some unusual archaeal viruses such as Ampullaviridae, Guttaviridae, Bicaudaviridae, Fuselloviridae, Thaspiviridae and Halspiviridae appear to be related to prokaryotic plasmids and transposons and indeed their genomes may have evolved from these non-viral mobile genetic elements. The families Ampullaviridae, Thaspiviridae and Halspiviridae show a close relationship with capson transposons, while the families Guttaviridae, Bicaudaviridae, and Fuselloviridae, are linked with some types of archaeal plasmids. The possibility has been raised that the enigmatic archaeal virosphere evolved by genetic recombination between non-viral mobile genetic elements and viruses. Also unassigned is the unusual bacterial DNA virus family Plasmaviridae, which also shows no phylogenetic relationship to other viruses and cellular organisms, whose provenance might be similar to that proposed for the unusual archaeal viruses.
It was formerly proposed that DNA viruses along with other DNA acellular ones should be grouped under the group Deoxyviria however apparently some DNA viruses are more closely related to cellular organisms (Cytota) than to other DNA viruses, making it paraphyletic.
Contenido relacionado
Lantana
Bladder tenesmus
Dinoflagellate