Bacterium
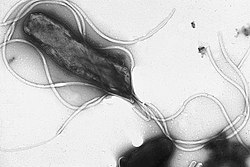
Bacteria are prokaryotic microorganisms that are a few micrometers in size (usually between 0.5 and 5 μm in length) and various shapes, including spheres (cocci), rods (bacilli), curved filaments (vibrios) and helical filaments (spirilla and spirochetes). Bacteria are prokaryotic cells, therefore, unlike eukaryotic cells (of animals, plants, fungi, etc.), they do not have a defined nucleus. nor do they present, in general, internal membranous organelles. They generally have a cell wall and this is made up of peptidoglycan (also called murein). Many bacteria have flagella or other locomotion systems to move. Bacteriology, a branch of microbiology, deals with the study of bacteria.
Although the term bacteria traditionally included all prokaryotes, taxonomy and scientific nomenclature currently divide them into two groups. These evolutionary domains are called Bacteria and Archaea (archaea). The division is justified by the great differences that both groups present at the biochemical and genetic level. The frequent presence of peptidoglycan wall together with its composition in membrane lipids are the main difference that they present compared to archaea.
Bacteria are the most abundant organisms on the planet. They are ubiquitous, found in all terrestrial and aquatic habitats; They grow even in the most extreme, such as in hot and acidic springs, in radioactive waste, in the depths of both the sea and the earth's crust. Some bacteria can even survive in the extreme conditions of outer space. It is estimated that around 40 million bacterial cells can be found in one gram of soil and one million bacterial cells in one milliliter of fresh water. In total, it is estimated that there are approximately 5×1030 bacteria in the world.
Bacteria are essential for the recycling of elements, since many important steps in biogeochemical cycles depend on them. Examples include the fixation of atmospheric nitrogen. However, only half of the known phyla of bacteria have species that can be cultivated in the laboratory, so a large part (estimated about 90%) of extant bacterial species have not yet been described.
In the human body there are approximately ten times as many bacterial cells as human cells, with a large number of bacteria on the skin and in the digestive tract. Although the protective effect of the immune system makes the vast majority of these bacteria harmless or beneficial, some pathogenic bacteria can cause infectious diseases, including cholera, diphtheria, scarlet fever, leprosy, syphilis, typhus, etc. The most common fatal bacterial diseases are respiratory infections, with a mortality for tuberculosis alone of about 1.5 million people in 2018. Antibiotics are used worldwide to treat bacterial infections. Antibiotics are effective against bacteria, as they inhibit the formation of the cell wall or stop other processes in their life cycle. They are also widely used in agriculture and livestock in the absence of disease, causing bacterial resistance to antibiotics to become widespread.
In industry, bacteria are important in processes such as wastewater treatment, in the production of butter, cheese, vinegar, yogurt, etc., and in the manufacture of medicines and other chemical products.
History of bacteriology
The existence of microorganisms was conjectured in the late Middle Ages. In the Canon of Medicine (1020), Abū Alī ibn Sīnā (Avicenna) stated that bodily secretions were contaminated by multiple infectious foreign bodies before a person fell ill., but he did not go so far as to identify these bodies as the first cause of the diseases. When the Black Death (bubonic plague) reached al-Andalus in the XIV century, Ibn Khatima and Ibn al-Khatib wrote that infectious diseases were caused by contagious entities penetrating the human body. These ideas about contagion as the cause of some diseases became very popular during the Renaissance, notably through the writings of Girolamo Fracastoro.
The first bacteria were observed by the Dutchman Anton van Leeuwenhoek in 1676 using a single-lens microscope designed by himself. He initially called them animalcules and published his observations in a series of letters he sent to to the Royal Society of London. Marc von Plenciz (18th century) stated that contagious diseases were caused by the small organisms discovered by Leeuwenhoek. The name bacteria was introduced later, in 1828, by Ehrenberg, derived from the Greek βακτήριον bacterion, meaning little stick. In 1835 Agostino Bassi, was able to demonstrate experimentally that the silkworm disease was of microbial origin, later deduced that many diseases such as typhus, syphilis and cholera would have a similar origin. In the classifications of the 1850s, bacteria with the name Schizomycetes were placed within the plant kingdom and in 1875 they were grouped together with blue-green algae in Schizophyta.
Louis Pasteur demonstrated in 1859 that fermentation processes were caused by the growth of microorganisms, and that said growth was not due to spontaneous generation, as was previously assumed. (Neither yeast, nor molds, nor fungi, organisms normally associated with these fermentation processes, are bacteria). Pasteur, like his contemporary and colleague Robert Koch, was an early proponent of the microbial theory of disease. Robert Koch was a pioneer in medical microbiology, working with a variety of infectious diseases, such as cholera, anthrax and tuberculosis. Koch managed to prove the microbial theory of disease after his research in tuberculosis, for which he was awarded the Nobel Prize in Medicine and Physiology in 1905. He established what has since been called the Koch's postulates, through which a series of experimental criteria were standardized to demonstrate whether or not an organism was the cause of a certain disease. These postulates are still used today.
Although at the end of the XIX century it was already known that bacteria were the cause of many diseases, there were no antibacterial treatments to combat them. In 1882 Paul Ehrlich, a pioneer in the use of dyes and dyes to detect and identify bacteria, discovered the Koch bacillus stain (Ziehl Neelsen stain) which was perfected shortly after by Ziehl and Neelsen independently. In 1884 Gram stain is found. Ehrlich received the Nobel Prize in 1908 for his work in the field of immunology and in 1910 developed the first antibiotic using dyes capable of selectively staining and killing spirochetes of the species Treponema pallidum, the bacterium that causes syphilis.
A breakthrough in the study of bacteria was the discovery by Carl Woese in 1977 that archaea have a different evolutionary lineage than bacteria. This new phylogenetic taxonomy was based on ribosomal RNA sequencing 16S and divided prokaryotes into two different evolutionary groups, in a system of three domains: Archaea, Bacteria, and Eukarya.
Origin and evolution of bacteria
Living beings are currently divided into three domains: bacteria (Bacteria), archaea (Archaea) and eukaryotes (Eukarya). The Archaea and Bacteria domains include prokaryotic organisms, that is, those whose cells do not have a differentiated cell nucleus, while the Eukarya domain includes the most well-known and complex forms of life (protists, animals, fungi, and plants)..
The term "bacteria" was traditionally applied to all prokaryotic microorganisms. However, molecular phylogeny has been able to show that prokaryotic microorganisms are divided into two domains, originally named Eubacteria and Archaebacteria, and now renamed Bacteria and Archaea, which evolved independently from a common ancestor. These two domains, together with the Eukarya domain, form the basis of the three-domain system, which is currently the most widely used classification system in bacteriology.
The term Monera, currently in disuse, in the ancient classification of the five kingdoms meant the same thing as prokaryote, and it continues to be used that way in many manuals and textbooks.
The ancestors of modern prokaryotes were the first organisms (the first cells) to develop on earth, more than 4.35 billion years ago. For about 3 billion more years, all organisms remained microscopic, with bacteria and archaea probably being the dominant life forms. Although bacterial fossils exist, for example stromatolites, because they do not retain their distinctive morphology they cannot be used for study the history of bacterial evolution, or the origin of a particular bacterial species. However, genetic sequences can be used to reconstruct the phylogeny of living things, and these studies suggest that archaea and eukaryotes are more related to each other than to bacteria.
It has been suggested that the last universal common ancestor of bacteria and archaea is a thermophile that lived 4.35 billion years ago during the Hadic. The bifurcation between archaea and bacteria occurred in the middle of the Hadic, while eukaryotes are more recent and emerged at the beginning of the Paleoproterozoic. Most of the bacterial phyla originated during the Archean. Thermophilic bacteria and ultrasmall bacteria (CPR) separated from the rest of the bacteria in the late Hadic and early Archean. The large bacterial clades Gracilicutes and Terrabacteria originated in the mid-Archaic.
It is currently debated whether the first prokaryotes were bacteria or archaea. Some researchers think that Bacteria is the older domain with Archaea and Eukarya deriving from it, while others consider Archaea to be the older domain. Instead, other scientists hold that both Archaea and Eukarya are relatively recent. (about 900 million years ago) and which evolved from a Gram-positive bacterium (probably an Actinobacteria), which by replacing the peptidoglycan bacterial wall with a glycoprotein wall would give rise to a Neomura organism.
Bacteria have also been implicated in the second great evolutionary divergence, the one that separated Archaea from Eukarya. Eukaryotic mitochondria are thought to have arisen from endosymbiosis of an alpha Proteobacterium. In this case, the eukaryotic ancestor, possibly related to archaea (the organism Neomura), ingested a proteobacterium that, escaping digestion, developed in the cytoplasm and gave rise to mitochondria. These can be found in all eukaryotes, although sometimes in very reduced forms, such as in mitochondrial protists. Later, and independently, a second endosymbiosis by some mitochondrial eukaryote with a cyanobacterium led to the formation of algae and plant chloroplasts. Some groups of algae are even known to have clearly originated from later events of endosymbiosis by heterotrophic eukaryotes that, after ingesting eukaryotic algae, became second-generation plastids.
Bacterial morphology
Bacteria come in a wide variety of sizes and shapes. Most have a size ten times smaller than that of eukaryotic cells, that is, between 0.5 and 5 μm. However, some species such as Thiomargarita namibiensis and Epulopiscium fishelsoni reach 0.5 mm and the largest known Thiomargarita magnifica reaches between 10 and 20 mm, which makes them visible to the naked eye. At the other extreme are the smallest known bacteria, including those belonging to the class Mollicutes, which includes the genus Mycoplasma and ultra-small bacteria (Patescibacteria or CPR group), which measure only 0.3 μm, that is, as small as the largest viruses.
The shape of bacteria is very varied and, often, the same species adopts different morphological types, which is known as pleomorphism. In any case, we can distinguish three fundamental types of bacteria:
- Coco (from the Greek kókkos, grain): spherical.
- Diploma: coconuts in groups of two.
- Tetraco: coconuts in groups of four.
- Estreptoco: coconuts in chains.
- Staph: coconuts in irregular or cluster groups.
- Bacilo (Latin baculus, rod): in the form of a stick.
- Helical forms:
- Vibrius: slightly curved and in the form of a comma, a Jew, peanut or arrinado.
- Sword: in rigid helical form or in the form of tyrant.
- Spirochete: in the form of a tyre (flex helical).
Some species even have tetrahedral or cubic shapes. This wide variety of shapes is ultimately determined by the composition of the cell wall and cytoskeleton, being of vital importance, since it can influence the ability of the bacterium to acquire nutrients, attach to surfaces, or move in the presence of stimuli.
The following are different species with different association patterns:
- Neisseria gonorrhoeae in diploid form (by pairs).
- Streptococcus chain-shaped.
- Staphylococcus in cluster form.
- Actinobacteria in the form of filaments. Such filaments tend to surround themselves with a pod that contains a multitude of individual cells, which can become branched, such as gender Nocardia, thus acquiring the appearance of the micelio of a fungus.
Bacteria have the ability to anchor themselves to certain surfaces and form a cell aggregate in the form of a layer called a biofilm or biofilm, which can have a thickness ranging from a few micrometers to half a meter. These biofilms can congregate various bacterial species, as well as protists and archaea, and are characterized by forming a conglomerate of cells and extracellular components, thus reaching a higher level of organization or secondary structure called microcolony, through in which there are many channels that facilitate the diffusion of nutrients. In natural environments such as soil or plant surfaces, most bacteria are anchored to surfaces in the form of biofilms. Said biofilms must be taken into account in chronic bacterial infections and in medical implants, since the bacteria that form these structures are much more difficult to eradicate than individual bacteria.
Finally, it is worth noting an even more complex type of morphology, observable in some microorganisms of the myxobacteria group. When these bacteria are in a medium low in amino acids, they are able to detect the surrounding cells, in a process known as quorum sensing, in which all cells migrate towards each other and aggregate, giving rise to fruiting bodies that they can reach 0.5 mm in length and contain about 100,000 cells. Once this structure is formed, the bacteria are capable of carrying out different functions, that is, they differentiate, thus reaching a certain level of multicellular organization. For example, between one and ten cells migrate to the upper part of the fruiting body and, once there, differentiate into a type of dormant cell called myxospores, which are more resistant to infection. desiccation and, in general, adverse environmental conditions.
Bacterial cell structure
Bacteria are relatively simple organisms. Its dimensions are very small, about 2 μm wide by 7-8 μm long in the cylindrical form (bacillus) of medium size; although species from 0.5 to 1.5 μm are very frequent.
As they are prokaryotic organisms, they have the corresponding basic characteristics such as the lack of a nucleus delimited by a membrane, although they present a nucleoid, an elementary structure that contains a large circular DNA molecule. The cytoplasm lacks organelles bounded by membranes and protoplasmic formations typical of eukaryotic cells. In the cytoplasm you can see plasmids, small circular DNA molecules that coexist with the nucleoid, contain genes and are commonly used by prokaryotes in conjugation. The cytoplasm also contains vacuoles (granules containing storage substances) and ribosomes (used in protein synthesis).
A cytoplasmic membrane made of lipids surrounds the cytoplasm, and like plant cells, most have a cell wall, which in this case is made of peptidoglycan (murein). Most bacteria also have a second lipid membrane (outer membrane) surrounding the cell wall. The space between the cytoplasmic membrane and the cell wall (or the outer membrane if it exists) is called the periplasmic space. Some bacteria present a capsule and others are capable of developing as endospores, latent states capable of withstanding extreme conditions. Among the external formations typical of the bacterial cell, the flagella and the pili stand out.
Intracellular Structures
The bacterial cytoplasmic membrane has a structure similar to that of plants and animals. It is a lipid bilayer composed mainly of phospholipids into which protein molecules are inserted. In bacteria it performs numerous functions including osmotic barrier, transport, biosynthesis, energy transduction, DNA replication center, and anchor point for flagella. Unlike eukaryotic membranes, it generally does not contain sterols (mycoplasmas and some proteobacteria are exceptions), although it may contain similar components called hopanoids.
Many important biochemical reactions that take place in cells are produced by the existence of concentration gradients on both sides of a membrane. This gradient creates a potential difference analogous to that of an electric battery and allows the cell, for example, to transport electrons and obtain energy. The absence of inner membranes in bacteria means that these reactions have to occur across the cytoplasmic membrane itself, between the cytoplasm and the periplasmic space.
Since bacteria are prokaryotes, they do not have membrane-bounded cytoplasmic organelles and therefore appear to have few intracellular structures. They lack a cell nucleus, mitochondria, chloroplasts, and the other organelles present in eukaryotic cells, such as the Golgi apparatus and endoplasmic reticulum. Some bacteria contain membrane-enclosed intracellular structures that can be considered primitive organelles, called prokaryotic compartments. Examples are the thylakoids of cyanobacteria, the ammonium monooxygenase-containing compartments in Nitrosomonadaceae, and various structures in Planctomycetes.
Like all living organisms, bacteria contain ribosomes for protein synthesis, but these are different from those of eukaryotes. The structure of ribosomes and ribosomal RNA of archaea and bacteria are similar, both ribosomes are of type 70S while eukaryotic ribosomes are of the 80S type. However, most ribosomal proteins, translation factors, and archaeal tRNAs are more eukaryotic-like than bacterial-like.
Many bacteria have vacuoles, intracellular granules for the storage of substances, such as glycogen, polyphosphates, sulfur or polyhydroxyalkanoates. Certain photosynthetic bacterial species, such as cyanobacteria, produce internal gas vesicles that they use to regulate their buoyancy and thus reach depth with optimal light intensity or optimal nutrient levels. Other structures present in certain species are carboxysomes (containing enzymes for carbon fixation) and magnetosomes (for magnetic orientation).

Bacteria do not have a membrane-bound nucleus. The genetic material is organized in a single chromosome located in the cytoplasm, within an irregular body called a nucleoid. Most bacterial chromosomes are circular, although there are some examples of linear chromosomes, for example, Borrelia burgdorferi. The nucleoid contains the chromosome along with associated proteins and RNA. The order Planctomycetes is an exception, as a membrane surrounds its nucleoid and has several membrane-bounded cell structures.
Prokaryotic cells were previously thought to have no cytoskeleton, but bacterial homologs of the major eukaryotic cytoskeletal proteins have since been found. These include the structural proteins FtsZ (which assembles into a ring to mediate during cell formation). bacterial cell division) and MreB (which determines the width of the cell). The bacterial cytoskeleton plays essential roles in protection, bacterial cell shape determination, and cell division.
Extracellular Structures
Bacteria have a cell wall that surrounds their cytoplasmic membrane. Bacterial cell walls are made of peptidoglycan (formerly called murein). This substance is composed of polysaccharide chains linked by unusual peptides that contain D amino acids. These amino acids are not found in proteins, so they protect the cell wall from most peptidases. Bacterial cell walls are different from those of plants and fungi, which are composed of cellulose and chitin, respectively. They are also different from the cell walls of Archaea, which do not contain peptidoglycan. The antibiotic penicillin can kill many bacteria by inhibiting a step in peptidoglycan synthesis.
There are two different types of bacterial cell wall called Gram-positive and Gram-negative. These names come from the reaction of the cell wall to Gram staining, a method traditionally used for the classification of bacterial species. Gram-positive bacteria have a thick cell wall that contains numerous layers of peptidoglycan into which it inserts. teichoic acid. In contrast, Gram-negative bacteria have a relatively thin wall, consisting of a few layers of peptidoglycan, surrounded by a second lipid membrane (the outer membrane) containing lipopolysaccharides and lipoproteins.
Mycoplasmas are an exception, as they lack a cell wall. Most bacteria have Gram-negative cell walls; only Firmicutes and Actinobacteria are Gram-positive. These two groups were formerly known as low GC Gram-positive bacteria and high GC Gram-positive bacteria, respectively. These differences in cell wall structure give rise to differences in antibiotic susceptibility. For example, vancomycin can only kill Gram-positive bacteria and is ineffective against Gram-negative pathogens, such as Haemophilus influenzae or Pseudomonas aeruginosa. Special mention must be made of the genus Mycobacterium, which, although it is classified as Gram positive, does not seem to be so from an empirical point of view, since its wall does not retain the dye. This is because they have an unusual cell wall, rich in mycolic acids, hydrophobic and waxy in nature and quite thick, which gives them great resistance.
Many bacteria have an S-layer of rigidly structured protein molecules that covers the cell wall. This layer provides chemical and physical protection for the cell surface and can act as a macromolecular diffusion barrier. The S layers have various (although not yet well understood) functions. For example, in the genus Campylobacter they act as virulence factors and in the species Bacillus stearothermophilus they contain surface enzymes.
Flagella are long filamentous appendages made of protein and used for movement. They are about 20 nm in diameter and up to 20 μm in length. The flagella are driven by energy obtained from ion transfer. This transfer is driven by the electrochemical gradient that exists between both sides of the cytoplasmic membrane.
Fimbriae are thin filaments of proteins that are distributed over the cell surface. They are approximately 2-10 nm in diameter and up to several μm in length. When viewed through an electron microscope, they resemble fine hairs. Fimbriae aid in the adherence of bacteria to solid surfaces or to other cells and are essential in the virulence of some pathogens. Pili are cellular appendages slightly larger than fimbriae and are used for the transfer of genetic material between bacteria in a process called bacterial conjugation.
Many bacteria are capable of accumulating material on the outside to cover their surface. Depending on the rigidity and their relationship with the cell, they are classified into capsules and glycocalyx. The capsule is a rigid structure that is firmly attached to the bacterial surface, while the glycocalyx is flexible and loosely attached. These structures protect bacteria by making it difficult for them to be phagocytosed by eukaryotic cells such as macrophages. They may also act as antigens and be involved in bacterial recognition, as well as aid in surface adhesion and biofilm formation.
The formation of these extracellular structures depends on the bacterial secretion system. This system transfers proteins from the cytoplasm to the periplasm or the space surrounding the cell. Many types of secretion systems, which are often essential for the virulence of pathogens, are known and are therefore extensively studied.
Endospores
- See also: Bacterial spores
Certain genera of Gram-positive bacteria, such as Bacillus, Clostridium, Sporohalobacter, Anaerobacter and Heliobacterium, can form endospores. Endospores are highly resistant, dormant structures whose primary function is to survive harsh environmental conditions. In almost all cases, endospores are not part of a reproductive process, although Anaerobacter can form up to seven endospores from one cell. Endospores have a central base of cytoplasm that contains DNA and ribosomes., surrounded by a bark and protected by a waterproof and rigid cover.
Endospores have no detectable metabolism and can survive extreme physical and chemical conditions, such as high levels of ultraviolet light, gamma rays, detergents, disinfectants, heat, pressure, and desiccation. In this dormant state, bacteria can continue to live for millions of years, and can even survive in the radiation and vacuum of outer space. Endospores can also cause disease. For example, anthrax can be contracted from inhalation of Bacillus anthracis endospores and tetanus from contamination of wounds with Clostridium tetani endospores.
Metabolism
In contrast to higher organisms, bacteria exhibit a wide variety of metabolic types. The distribution of these metabolic types within a group of bacteria has traditionally been used to define their taxonomy, but these traits often do not correspond with modern genetic classifications. Bacterial metabolism is classified based on three important criteria: carbon origin, energy source, and electron donors. An additional criterion for classifying respiring microorganisms is the electron acceptor used in respiration.
Based on the carbon source, bacteria can be classified as:
- HeterotrophasWhen they use organic compounds.
- Autrophas, when cell carbon is obtained by fixing carbon dioxide.
Typical autotrophic bacteria are photosynthetic cyanobacteria, green sulfur bacteria, and some purple bacteria. But there are also many other chemolithotrophic species, for example, nitrifying and sulfur-oxidizing bacteria.
Depending on the energy source, bacteria can be:
- Phototrofaswhen they use light through photosynthesis.
- Chemotrophas, when they get energy from chemicals that are oxidized mainly at the expense of oxygen (aerobic breath) or other electron receptors (anaerobic breath).
Based on electron donors, bacteria can also be classified as:
- Litotrophas, if used as inorganic compound electron donors.
- Organizational, if used as donators of organic compound electrons.
Chemotrophs use electron donors for energy conservation (during aerobic and anaerobic respiration, and fermentation) and for biosynthetic reactions (for example, for carbon dioxide fixation), whereas phototrophs use them. They are used for biosynthetic purposes only.
Respiring organisms use chemical compounds as an energy source, taking electrons from the reduced substrate and transferring them to a terminal electron acceptor in a redox reaction. This reaction releases energy that can be used to synthesize ATP and thus keep the metabolism active. In aerobic organisms, oxygen is used as an electron acceptor. In anaerobic organisms other inorganic compounds such as nitrates, sulfates or carbon dioxide are used as electron acceptors. This leads to the important biogeochemical processes of denitrification, sulfate reduction, and acetogenesis, respectively, taking place. Another possibility is fermentation, an incomplete oxidation process, totally anaerobic, with the final product being an organic compound, which when reduced will be the final receptor of the electrons. Examples of reduced fermentation products are lactate (in lactic fermentation), ethanol (in alcoholic fermentation), hydrogen, butyrate, etc. Fermentation is possible because the energy content of the substrates is higher than that of the products, allowing organisms to synthesize ATP and keep their metabolism active. Facultative anaerobic organisms can choose between fermentation and various receptor terminals of electrons depending on the environmental conditions in which they are found.
Lithotrophic bacteria can use inorganic compounds as a source of energy. The most common inorganic electron donors are hydrogen, carbon monoxide, ammonia (leading to nitrification), ferrous iron and other reduced metal ions, as well as various reduced sulfur compounds. On certain occasions, methanotrophic bacteria can use methane gas as an electron source and substrate simultaneously, for carbon anabolism. In aerobic phototrophy and chemolithotrophy, oxygen is used as the terminal electron acceptor, while under anaerobic conditions it is use inorganic compounds. Most lithotrophic organisms are autotrophic while organotrophic organisms are heterotrophic.
In addition to fixing carbon dioxide through photosynthesis, some bacteria also fix nitrogen gas using the enzyme nitrogenase. This characteristic is very important at the environmental level and can be found in bacteria of almost all the metabolic types listed above, although it is not universal. Microbial metabolism can play an important role in bioremediation as, for example, some species can perform the wastewater treatment and others are capable of degrading hydrocarbons, toxic and even radioactive substances. Instead, sulfate-reducing bacteria are largely responsible for the production of highly toxic forms of mercury (methyl- and dimethyl-mercury) in the environment.
Movement
Some bacteria are immobile and others limit their movement to changes in depth. For example, cyanobacteria and green sulfur bacteria contain gas vesicles with which they can control their buoyancy and thus achieve optimal light and food. Motile bacteria can move by gliding, twitching, or more commonly using flagella. Some bacteria can slide down solid surfaces secreting a viscous substance, but the mechanism that acts as a propellant is still unknown. In contraction movement, the bacterium uses its type IV pilus as a grappling hook, first extending it, anchoring it, and then contracting it with remarkable force (>80 pN).
The bacterial flagellum is a long, filamentous, helical appendage propelled by a rotating motor (like a propeller) that can rotate in both directions. The motor uses as energy an electrochemical gradient across the membrane. Flagella are made up of about 20 proteins, with about 30 other proteins for their regulation and coordination. It must be taken into account that, given the size of the bacterium, water is very viscous for them and the propulsion mechanism must be very powerful and efficient. Bacterial flagella are found in both Gram-positive and Gram-negative bacteria and are completely different from eukaryotic flagella and, although superficially similar to archaeal ones, are considered non-homologous.

Depending on the number and arrangement of the flagella on the surface of the bacterium, the following types are distinguished: a single flagellum (monothrichous), a flagellum at each end (amphitric), groups of flagella at one or both ends (lophotricho) and flagella distributed over the entire cell surface (perithorico). In a unique group of bacteria, the spirochetes, there are specialized flagella, called axial filaments, located intracellularly in the periplasmic space, between the two membranes. These produce a rotary movement that causes the bacteria to rotate like a corkscrew moving forward.
Many bacteria (such as E. coli) have two types of movement: straight line (running) and random. In the latter, a random three-dimensional movement is performed by combining the bacterium with sprints and random turns. Motile bacteria can exhibit attractive or repulsive movements determined by different stimuli. These behaviors are called taxis, and include various types such as chemotaxis, phototaxis, or magnetotaxis. In the peculiar group of myxobacteria, individual cells move together to form waves of cells, which will eventually aggregating to form the characteristic fruiting bodies of this genus. Movement of myxobacteria occurs only on solid surfaces, in contrast to E. coli, which is mobile in both liquid and solid media.
Various species of Listeria and Shigella move within host cells by appropriating their cytoskeleton, which would normally move organelles. Actin polymerization creates a push on one end of the bacterium that moves it through the cytoplasm of the host cell.
The described mechanisms of bacterial movement detail how bacteria move spatially on a microscopic scale. The movement of bacteria on a larger scale is often referred to as "dispersal" and depends on passive mechanisms. In the airborne environment, the bacteria are transmitted attached to larger particles. Previous studies show how most bacteria are found in the fraction of air that contains particles larger than 10µm; these works show a mechanism by which pollen could be the main bacterial vector through the air.
Playback
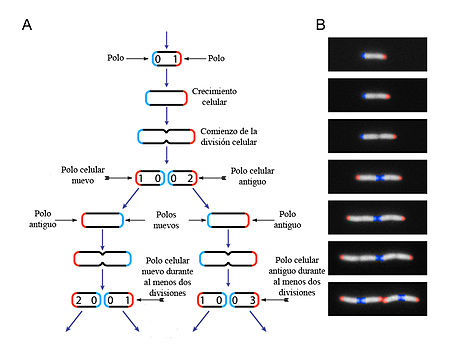
In bacteria, cell increase in size (growth) and reproduction by cell division are closely linked, as in most single-celled organisms. It occurs by duplication and cells with identical hereditary information are obtained. Bacteria grow to a fixed size and then reproduce by binary fission, a form of asexual reproduction. Under appropriate conditions, a Gram-positive bacterium can divide every 20–30 minutes and a Gram-negative bacterium every 15–20 minutes, and in about 16 hours their number can rise to about 5 billion (close to the number of people on Earth, which is approximately 7 billion people). Under optimal conditions, some bacteria can grow and divide very quickly, as much as every 9.8 minutes. In cell division, two identical daughter cells are produced. Some bacteria, still reproducing asexually, form more complex reproductive structures that facilitate the dispersal of newly formed daughter cells. Examples include the formation of fruiting bodies (sporangia) in myxobacteria, the formation of hyphae in Streptomyces, and budding. In budding a cell forms a bulge which then separates and produces a new daughter cell.
On the other hand, it is worth noting a type of sexual reproduction in bacteria, called bacterial parasexuality. In this case, the bacteria are capable of exchanging genetic material in a process known as bacterial conjugation. During the process, a donor bacterium and a recipient bacterium make contact through hollow sexual hairs or pili, through which a small amount of independent DNA or conjugative plasmid is transferred. The best known is the F plasmid of E. coli, which can also integrate into the bacterial chromosome. In this case it is called an episome, and in the transfer it carries part of the bacterial chromosome. DNA synthesis is required for conjugation to occur. The replication is done at the same time as the transfer.
Growth
Bacterial growth follows three phases. When a bacterial population finds itself in a new environment with a high concentration of nutrients that allow it to grow, it needs a period of adaptation to said environment. This first phase is called the adaptation phase or lag phase and involves slow growth, where cells prepare to begin rapid growth, and a high rate of biosynthesis of the proteins necessary for this, such as ribosomes., membrane proteins, etc. The second phase of growth is called the exponential phase, as it is characterized by the exponential growth of cells. The rate of growth during this phase is known as the growth rate k and the time it takes for each cell to divide as the generation time g. During this phase, nutrients are metabolized at the highest possible rate, until these nutrients are exhausted, giving way to the next phase. The last phase of growth is called the stationary phase and occurs as a consequence of the depletion of nutrients in the medium. In this phase, cells drastically reduce their metabolic activity and begin to use non-essential cellular proteins as an energy source. The stationary phase is a transition period from rapid growth to a state of response to stress, in which the expression of genes involved in DNA repair, antioxidant metabolism, and nutrient transport is activated.
Genetics

Most bacteria have a single circular chromosome whose size can range from just 160,000 base pairs in the endosymbiont bacterium Candidatus Carsonella ruddii to 12,200,000 base pairs in the bacterium of the endosymbiont. soil Sorangium cellulosum. Spirochetes of the genus Borrelia (including, for example, Borrelia burgdorferi, the cause of Lyme disease) they are a notable exception to this rule as they contain a linear chromosome. Bacteria may also have plasmids, small extra-chromosomal DNA molecules that may contain genes responsible for antibiotic resistance or virulence factors. Another type of bacterial DNA comes from the integration of genetic material from bacteriophages (viruses that infect bacteria). There are many types of bacteriophages, some simply infect and rupture bacterial host cells, while others insert into the bacterial chromosome. In this way, virus genes that contribute to the phenotype of the bacteria can be inserted. For example, in the evolution of Escherichia coli O157:H7 and Clostridium botulinum, toxic genes contributed by a bacteriophage turned a harmless ancestral bacterium into a lethal pathogen.
Bacteria, as asexual organisms, inherit identical copies of genes, that is, they are clones. However, they can evolve by natural selection through DNA changes due to mutations and genetic recombination. Mutations result from errors during DNA replication or from exposure to mutagenic agents. Mutation rates vary widely between different species of bacteria and even between different strains of the same species of bacteria. Genetic changes can occur randomly or be selected by stress, where genes involved in some process that limits growth they have a higher mutation rate.
Bacteria can also transfer genetic material between cells. This can be done in three main ways. First, bacteria can pick up foreign DNA from the environment in a process called transformation. Genes can also be transferred by a process of transduction whereby a bacteriophage introduces foreign DNA into the bacterial chromosome. The third method of gene transfer is by bacterial conjugation, where DNA is transferred through direct contact (via a pilus) between cells. This acquisition of genes from other bacteria or from the environment is called horizontal gene transfer and may be common under natural conditions. Gene transfer is especially important in antibiotic resistance, as it allows rapid dissemination of the genes responsible for such resistance. between different pathogens.
Interactions with other organisms
Despite their apparent simplicity, bacteria can form complex associations with other organisms. These associations can be classified as parasitism, mutualism, and commensalism.
Diners
Because of their small size, commensal bacteria are ubiquitous and grow on animals and plants just as they would on any other surface. Thus, for example, large populations of these organisms are the cause of bad body odor and their growth can be increased by heat and sweat.
Mutualists
Certain bacteria form intimate associations with other organisms, which are essential for their survival. One of these mutualistic associations is the transfer of hydrogen between species. It occurs between groups of anaerobic bacteria that consume organic acids such as butyric acid or propionic acid and produce hydrogen, and the methanogenous archaea that consume that hydrogen. Bacteria in this association cannot consume organic acids when hydrogen accumulates around them. around. Only close association with the archaea maintains a low enough hydrogen concentration to allow bacteria to grow.
In the soil, microorganisms that inhabit the rhizosphere (the area that includes the root surface and the soil that adheres to it) carry out nitrogen fixation, converting atmospheric nitrogen (in a gaseous state) into nitrogenous compounds This provides many plants, which cannot fix nitrogen on their own, with an easily absorbable form of nitrogen.
Many other bacteria are found as symbionts in humans and other organisms. For example, about a thousand bacterial species proliferate in the digestive tract. They synthesize vitamins such as folic acid, vitamin K and biotin. They also ferment indigestible complex carbohydrates and convert milk sugars into lactic acid (e.g. Lactobacillus). In addition, the presence of this intestinal flora inhibits the growth of potentially pathogenic bacteria (usually by competitive exclusion). Many times these beneficial bacteria are sold as probiotic dietary supplements.
Pathogens
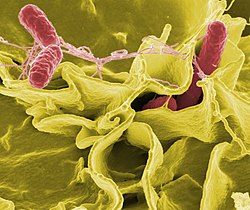
Only a small fraction of bacteria cause disease in humans: of the 15,919 species recorded in the NCBI database, only 538 are pathogenic. Yet they are a leading cause of human disease and mortality, causing infections such as tetanus, typhoid fever, diphtheria, syphilis, cholera, food poisoning, leprosy and tuberculosis. There are cases in which the etiology or cause of a known disease is discovered only after many years, as was the case with peptic ulcer and Helicobacter pylori. Bacterial diseases are also important in agriculture and livestock, where there are many diseases such as leaf spot, fire plague, paratuberculosis, bacterial panicle blight, mastitis, salmonella and anthrax.
Each species of pathogen has a characteristic spectrum of interactions with its human hosts. Some organisms, such as Staphylococcus or Streptococcus, can cause skin infections, pneumonia, meningitis, and even sepsis, a systemic inflammatory response that produces shock, massive vasodilatation, and death. However, these organisms are also part of the normal human flora and are usually found on the skin or in the nose without causing any disease.
Other organisms invariably cause disease in humans. For example, the genus Rickettsia, which are obligate intracellular parasites capable of growing and reproducing only within the cells of other organisms. One species of Rickettsia causes typhus, while another causes Rocky Mountain fever. Chlamydiae, another phylum of obligate intracellular parasites, contains species that cause pneumonia, urinary infections, and may be implicated in coronary heart disease. Finally, certain species such as Pseudomonas aeruginosa, Burkholderia cenocepacia i> and Mycobacterium avium are opportunistic pathogens and cause disease primarily in people with immunosuppression or cystic fibrosis.
Bacterial infections can be treated with antibiotics, which are classified as bactericidal, if they kill bacteria, or bacteriostatic, if they only stop bacterial growth. There are many types of antibiotics, and each type inhibits a process that differs in the pathogen from the host. Examples of selectively toxic antibiotics are chloramphenicol and puromycin, which inhibit the bacterial ribosome, but not the structurally different eukaryotic ribosome. The antibiotics are used to treat human diseases and in factory farming to promote animal growth. The latter may contribute to the rapid development of antibiotic resistance in bacterial populations. Infections can be prevented with antiseptic measures such as sterilizing the skin before injections and proper care of catheters. Surgical and dental instruments are also sterilized to prevent bacterial contamination and infection. Disinfectants such as bleach are used to kill bacteria or other pathogens that settle on surfaces to prevent contamination and reduce the risk of infection.
The following table shows some human diseases caused by bacteria:
| Disease | Agent | Main symptoms |
|---|---|---|
| Brucelosis | Brucella spp. | Worthy fever, adenopathy, endocarditis, pneumonia. |
| Carbunco | Bacillus anthracis | Fever, skin papula, septicemia. |
| Cholera | Vibrio cholerae | Diarrhea, vomiting, dehydration. |
| Diphtheria | Corynebacterium diphtheriae | Fever, tonsillitis, throat membrane, skin lesions. |
| Escarlatina | Streptococcus pyogenes | Fever, tonsillitis, erythema. |
| Erisipela | Streptococcus spp. | Fever, erythema, pruritus, pain. |
| Fiber Q | Coxiella burnetii | High fever, intense headache, myalgia, confusion, vomiting, diarrhea. |
| Typhoid fever | Salmonella typhi, S. paratyphi | High fever, bacteriemia, cefalalgia, stupor, nasal mucosa tumefaction, toasted tongue, ulcers in the palate, hepatoesplenomegaly, diarrhea, intestinal perforation. |
| Legionelosis | Legionella pneumophila | Fever, pneumonia |
| Pneumonia | Streptococcus pneumoniae, Staphylococcus aureus, Klebsiella pneumoniae, Mycoplasma spp., Chlamydia spp. | High fever, yellowish or bloody expectoration, chest pain. |
| Tuberculosis | Mycobacterium tuberculosis | Fever, tiredness, night sweat, lung necrosis. |
| Tetanus | Clostridium tetani | Fever, paralysis. |
Classification and identification
Taxonomic classification seeks to describe and differentiate the wide diversity of bacterial species by naming and grouping organisms according to their similarities. Bacteria can be classified based on different criteria, such as cell structure, metabolism, or based on differences in certain components such as DNA, fatty acids, pigments, antigens, or quinones. However, although these criteria allowed the identification and classification of bacterial strains, it was not yet clear whether these differences represented variations between different species or between different strains of the same species. This uncertainty was due to the absence of distinctive structures in most bacteria and the existence of horizontal gene transfer between different species, which means that closely related bacteria can have very different morphologies and metabolisms. For this reason, and in order to overcome this uncertainty, the current bacterial classification focuses on the use of modern molecular techniques (molecular phylogeny), such as the determination of guanine/cytosine content, genome-genome hybridization or sequencing of ribosomal DNA, which is not involved in horizontal transfer.
The International Committee for the Systematics of Prokaryotes (ICSP) is the body responsible for the nomenclature, taxonomy, and standards by which prokaryotes are designated. The ICSP is responsible for the publication of the International Code of Bacterial Nomenclature (list of approved names of bacterial species and taxa). It also publishes the International Journal of Systematic Bacteriology. In contrast to prokaryotic nomenclature, there is no official classification of prokaryotes because taxonomy remains a matter. of scientific criteria. The most accepted classification is the one elaborated by the editorial office of Bergey's Manual of Systematic Bacteriology (Bergey's Manual of Systematic Bacteriology) as a preliminary step to organize the content of the publication. This classification, known as " The Taxonomic Outline of Bacteria and Archaea" (TOBA), is available on the Internet. Due to the recent introduction of molecular phylogeny and genome sequence analysis, bacterial classification today is a constantly changing and expanding field.
The identification of bacteria in the laboratory is particularly relevant in medicine, where the determination of the species causing an infection is crucial when it comes to applying the correct treatment. Therefore, the need to identify human pathogens has led to a powerful development of techniques for the identification of bacteria.
The Gram staining technique for bacterial membranes, developed by Hans Christian Gram in 1884, has marked a before and after in the field of medicine, and consists of staining various bacterial samples with specific dyes in a slides to know if they have been stained or not with said dye.
Once the specific dyes have been added to the samples, and the sample has been washed after a few minutes to avoid confusion, they must be cleaned with a few drops of ethyl alcohol. The function of alcohol is to remove the stain from the bacteria, and this is where the bacteria that have been taken are recognized: if the bacteria retain the stain, it is a Gram positive, which have a thicker wall made up of several dozen of layers of various protein components; in the event that the stain does not hold, the bacterium is a Gram negative, which has a wall of a different composition. The biological function of this technique is to manufacture specific antibiotics for these bacteria.
This stain is used in microbiology to visualize bacteria in clinical samples. It is also used as a first step in the distinction of different species of bacteria, considering Gram positive bacteria to those that turn purple and Gram negative to those that turn red.
In the analysis of clinical samples it is usually a fundamental study because it fulfills several functions:
- Preliminary identification of the bacteria causing the infection.
- Considering the quality of the biological sample for the study, that is, it allows to appreciate the number of inflammatory cells as well as epithelial cells. The greater number of inflammatory cells in each microscope field, the more likely the growing flora in the culture media to be the representative of the infected area. A greater number of epithelial cells happen the opposite, greater probability of contamination with saprofita flora.
- Utility as quality control of bacterial isolation. Bacterial strains identified in the Gram stain should correspond to bacterial isolations made in the crops. If more bacterial forms are observed than isolated ones, then the means of used crops and the incubation atmosphere must be reconsidered.
Phyla and phylogeny
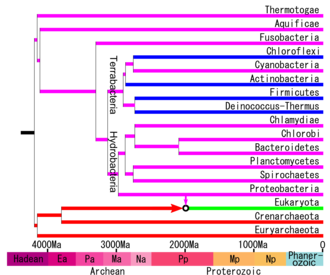
The phylogenetic relationships of living beings are controversial and there is no general agreement among different authors. Most phylogenetic trees, especially those of 16S and 23S rRNA, show that the basal groups are thermophilic phyla such as Aquificae and Thermotogae, which would reinforce the thermophilic origin of the Archaea and Bacteria domains. Instead, some genomic trees show Firmicutes (Gram positive) as the oldest clade. According to Cavalier-Smith theories the greatest divergence is in a photosynthetic group they call Chlorobacteria (Chloroflexi). Other genomic or protein phylogenetic studies place Planctomycetes, Proteobacteria, or other phyla in a basal position. Finally, it has been proposed that there was an early divergence between two supergroups: Gracilicutes and Terrabacteria; in short, demonstrating that there is currently no stable bacterial phylogeny to know with certainty the earliest bacterial evolutionary history. This is probably due to the phenomenon of horizontal gene transfer, typical of prokaryotic organisms.
The main bacterial phyla can be organized within a broad phylogenetic criteria into three sets:
Thermophilic groups
According to most molecular phylogenetic trees, thermophilic bacteria are the most divergent, forming a basal paraphyletic group, which is compatible with the main theories of prokaryotic origin and evolution. They are thermophiles and hyperthermophiles with chemotrophic metabolism, anaerobic respiration and Gram negative structure (double membrane), highlighting the following phyla:
- Take it. Small group of chemotherapy, thermophiles, or hyperthermal bacteria. It is found in hot springs, sulfurous wells and oceanic hydrothermal sources.
- Thermotogae. A phyll of hyperthermophiles, forced anaerobics, fermentative heterotrophos.
- Dictyoglomi. It includes a single species of hyperthermophile, chemoorganotropho and aerobic.
- Thermodesulfobacteria: Sulfate reducer thermophiles.
- Caldiserica. Anaerobic thermophile.
- Synergistetes. Anaerobic bacteria. Although few genera are thermophiles, Synergistetes has a basal position on the bacterial phylogen of RNA.
Gram positive and related
The Gram positive groups are basically Firmicutes and Actinobacteria, which would have thickened their cell wall as an adaptation to desiccation with loss of the outer membrane, developing sterols, teichoic acid and forming spores in several groups. The term Posibacteria has been used as a taxon to group Gram positives and derived groups such as Tenericutes. The term monodermic refers to the unique cell membrane that Gram positives have, which means that other phyla such as Chloroflexi and Thermomicrobia, being monodermic, are related to the former despite being Gram variable. According to some phylogenetic trees, the monodermic phyla form part of a superclade called Terrabacteria, named for their probable evolution in terrestrial environments, and included didermic phyla such as Deinococcus-Thermus, which is Gram variable and to the Cyanobacteria/Melainabacteria group which is Gram negative. Gram-positive and related (Terrabacteria) occur in most phylogenetic trees as a paraphyletic group with respect to Gracilicutes and is made up of the following phyla:
- Actinobacteria. An extensive edge of positive Gram bacteria of high GC content. They are common on the ground although some inhabit plants and animals, including some pathogens. Some form colonies in the form of fifas (Actinomyces).
- Firmicutes or Endobacteria. It is the largest group and includes positive Gram bacteria with low GC content. They are found in various habitats, including some notable pathogens. One of the families, Heliobacteria, gets its energy through photosynthesis and others have a pseudo outer membrane (Negativicutes).
- Tenericutes or Mollicutes. They are endosimbionts Monodermal negative gram and no cell wall. They are derived from Firmicutes according to most philgenies.
- Deinococcus-Thermus or Hadobacteria. Small group of highly resistant quimiorganotrophos. Some species support extreme heat and cold, while others are resistant to radiation and toxic substances.
- Chloroflexi. Small edge of monodermal bacteria Gram optional and typically filamentous aerobic variables. It includes non-sulphur green bacteria, which perform anoxygenic photosynthesis through bacteriochlorophyll (without oxygen production) and where their carbon fixation path also differs from that of other photoynthetic bacteria.
- Thermomicrobia. Small thermophile (or class) derived from Chloroflexi.
- Cyanobacteria (green-blue algae). The most important group of photosynthetic bacteria. They present chlorophyll and perform oxygenic photosynthesis. They are unicellular or colonial filaments.
- Armatimonadetes. Small aerobic chemoheterotrophic group.
Gracilicutes
The superclade Gracilicutes or Hydrobacteria is well agreed upon in many phylogenetic trees. They are the largest group of Gram-negative, didermal, mostly chemoheterotrophic bacteria, from aquatic habitats or related to animals and plants as a commensal, mutualist, or pathogen. It is made up of several phyla and superphyla:
- Spirochaetes. Chemoheterotrophaetic bacteria with elongated shape typically coiled in spiral that move by rotation. Many produce diseases.
- FCB or Sphingobacteria Group.
- Fibrobacters. Small edge that includes many of the stomach bacteria that allow cellulose degradation in ruminants.
- Gemmatimonadetes. Floor aerobics and mud.
- Bacteroids. An extensive edge of bacteria with wide distribution in the environment, including soil, sediment, sea water and the digestive tract of animals. It is a heterogeneous group that includes forced aerobics or forced anaerobics, comenals, parasites and free life forms.
- Chlorobi. They highlight the green sulfur bacteria, which are phototrophas through bacteriochlorophyll and forced anaerobics. Some are thermophiles that live in hydrothermal sources. They are considered derivatives or related to phyll-level Bacteroidetes.
- PVC or Planctobacteria
- Planctomycetes. Bacteria mainly aerobic aerobics found in fresh, salt and marine water. Its biological cycle involves the alternation between sesile and flogged cells. They reproduce by gem.
- Verrucomicrobia. It includes terrestrial, aquatic bacteria and some associated with eukaryotic guests.
- Lentisphaerae. Small group of newly discovered bacteria in marine waters and anaerobic land habitats. They are considered derivatives or related to Verrucomicrobia at the level of phyll.
- Chlamydiae. A small group of forced intracellular parasites of eukaryotic cells.
- Elusimicrobia. It is scattered by sea, land and as endosimbionte of insects.
- Proteobacteria (purple and related bacteria). It is a very diverse and extensive group. Most are heterotrophas, others are fermenters like enterobacteria and many cause diseases such as intracellular parasites. Rhizobes are endosimbionts of nitrogen fixers in plants, purple bacteria are phototrophas with bacteriochlorophyll and mixedobacteria form multicellular aggregates. Some authors consider that the following are derivatives or related to Proteobacteria:
- Acidobacteria. Small edge of common acidophyll bacteria in the soil. It includes a phototropha bacteria using bacteriochlorophyll.
- Deferribacters. Small group of anaerobic aquatic bacteria.
- Chrysiogenetes. Small anaerobic chemotherapeutic group with a unique biochemistry and lifestyle, capable of breathing arseniato.
- Nitrospirae. Group of nitrogen oxidant chemosynthetics and some are thermophiles.
- Fusobacteria. It's not always included in Gracilicutes. They are anaerobic heterotrophae bacteria that cause human infections. They constitute one of the main types of flora of the digestive system.
CPR group and other candidate phyla
Recently, genomic analyzes of samples taken from the environment have identified a large number of candidate phyla of ultrasmall bacteria, whose representatives have not yet been cultivated. These bacteria had not been detected by traditional procedures due to their special metabolic characteristics. As an example, a new phylogenetic line of bacteria containing 35 phyla, the CPR group, has recently been identified. In this way, the number of phyla of the Bacteria domain is extended to almost 100 and far exceeds in diversity the organisms of the other two domains. However, in 2018 in a taxonomic review to standardize bacterial taxonomy it was found that CPR or Patescibacteria comprises a single phylum with several lower-rank taxa, rather than a radiation of multiple phyla.
Use of bacteria in technology and industry
Many industries depend in part or entirely on bacterial action. A large number of important chemicals such as ethyl alcohol, acetic acid, butyl alcohol and acetone are produced by specific bacteria. Bacteria are also used for curing tobacco, leather, rubber, cotton, etc. Bacteria (often Lactobacillus) along with yeasts and molds, have been used for thousands of years for the preparation of fermented foods such as cheese, butter, pickles, soy sauce, sauerkraut, vinegar, wine and yogurt.
Bacteria have a remarkable ability to degrade a wide variety of organic compounds, which is why they are used in garbage recycling and bioremediation. Bacteria capable of breaking down hydrocarbons are often used in cleaning up oil spills. For example, after the Exxon Valdez oil spill in 1989, fertilizers were used on some beaches in Alaska to promote the growth of oil. these natural bacteria. These efforts were effective on beaches where the oil layer was not too thick. Bacteria are also used for bioremediation of industrial toxic wastes. In the chemical industry, bacteria are used in the synthesis of enantiomerically pure chemicals for pharmaceutical or agrochemical use.
Bacteria can also be used for biological parasite control instead of pesticides. This commonly involves the species Bacillus thuringiensis (also called BT), a Gram-positive soil bacterium. Subspecies of this bacterium are used as Lepidopteran-specific insecticides. Due to their specificity, these pesticides are considered environmentally friendly, with little or no effect on humans, wildlife, and most beneficial insects, such as for example, pollinators.
Bacteria are basic tools in the fields of molecular biology, genetics, and biochemistry due to their ability to grow rapidly and the relative ease with which they can be manipulated. By making modifications to bacterial DNA and examining the resulting phenotypes, scientists can determine the function of genes, enzymes, and metabolic pathways, and can later translate this knowledge to more complex organisms. Understanding cellular biochemistry, which requires enormous amounts of data related to enzyme kinetics and gene expression will allow mathematical models of entire organisms. This is feasible in some well-studied bacteria. For example, the Escherichia coli metabolism model is currently being developed and tested. This understanding of bacterial metabolism and genetics allows biotechnology to modify bacteria to produce various therapeutic proteins, such as insulin, growth factors and antibodies.
Gallery
Contenido relacionado
Princess of Asturias Award for Scientific and Technical Research
Carlos Noriega
Metamer